The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation
- PMID: 18498745
- PMCID: PMC2633441
- DOI: "V体育安卓版" 10.1016/j.molcel.2008.03.009
V体育官网入口 - The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation
V体育ios版 - Abstract
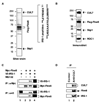
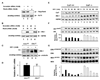
Recent genetic studies have documented a pivotal growth-regulatory role played by the Cullin 7 (CUL7) E3 ubiquitin ligase complex containing the Fbw8-substrate-targeting subunit, Skp1, and the ROC1 RING finger protein. In this report, we identified insulin receptor substrate 1 (IRS-1), a critical mediator of the insulin/insulin-like growth factor 1 signaling, as a proteolytic target of the CUL7 E3 ligase in a manner that depends on mammalian target of rapamycin and the p70 S6 kinase activities. Interestingly, while embryonic fibroblasts of Cul7-/- mice were found to accumulate IRS-1 and exhibit increased activation of IRS-1's downstream Akt and MEK/ERK pathways, these null cells grew poorly and displayed phenotypes reminiscent of those associated with oncogene-induced senescence. Taken together, our findings demonstrate a key role for the CUL7 E3 in targeting IRS-1 for degradation, a process that may contribute to the regulation of cellular senescence VSports手机版. .
"V体育ios版" Figures





References
-
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995;270:27489–27494. - PubMed
-
- Andrews P, He YJ, Xiong Y. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antagonizing p53 function. Oncogene. 2006;25:4534–4548. - PubMed
-
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. - PubMed
-
- Daud AI, Lanson NA, Jr, Claycomb WC, Field LJ. Identification of SV40 large T-antigen-associated proteins in cardiomyocytes from transgenic mice. Am. J. Physiol. 1993;264:H1693–H1700. - "V体育2025版" PubMed
Publication types (V体育官网入口)
- Actions (VSports最新版本)
MeSH terms
- "V体育平台登录" Actions
- Actions (V体育2025版)
- "VSports最新版本" Actions
- VSports注册入口 - Actions
- "VSports最新版本" Actions
- V体育2025版 - Actions
- V体育ios版 - Actions
- V体育ios版 - Actions
- "VSports注册入口" Actions
- V体育官网入口 - Actions
- "V体育官网" Actions
- Actions (V体育官网)
- V体育平台登录 - Actions
- VSports注册入口 - Actions
Substances
- "VSports在线直播" Actions
- Actions (VSports)
- "V体育2025版" Actions
- Actions (VSports最新版本)
- "VSports注册入口" Actions
- VSports最新版本 - Actions
- VSports app下载 - Actions
- VSports手机版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

