Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain
- PMID: 18474648
- PMCID: PMC2446731
- DOI: 10.1128/IAI.00366-08
Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain (VSports)
Abstract
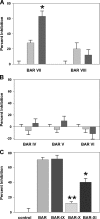
Porphyromonas gingivalis initially colonizes the oral cavity by interacting with organisms in supragingival plaque, such as the oralis group of oral streptococci. This interaction involves the association of the streptococcal antigen I/II with the minor fimbrial antigen (Mfa1) of P. gingivalis. Our previous studies showed that a peptide (BAR) derived from antigen I/II inhibits P. gingivalis adherence and subsequent biofilm formation on streptococcal substrates. In addition, screening a combinatorial peptide library identified select amino acid substitutions in the NITVK active region of BAR that increased the adherence of P. gingivalis to streptococci. Here we report that incorporating these residues in a synthetic peptide results in more-potent inhibition of P. gingivalis adherence and biofilm formation (I(50) [50% inhibition] at 0 VSports手机版. 52 microM versus I(50) at 1. 25 microM for BAR). In addition, a second structural motif in BAR, comprised of the amino acids KKVQDLLKK, was shown to contribute to P. gingivalis adherence to streptococci. Consistent with this, the KKVQDLLKK and NITVK motifs are conserved only in antigen I/II proteins expressed by the oralis group of streptococci, which interact with P. gingivalis. Interestingly, the primary and secondary structures and the functional characteristics of the amphipathic VQDLL core alpha-helix resemble the consensus nuclear receptor (NR) box protein-protein interacting domain sequence (LXXLL) of eukaryotes. BAR peptides containing amino acid substitutions with the potential to disrupt the secondary structure of VQDLL were less-effective inhibitors of P. gingivalis adherence and biofilm formation, suggesting that the alpha-helical character of VQDLL is important. Furthermore, replacing the lysines that flank VQDLL with acidic amino acids also reduced inhibitory activity, suggesting that the association of VQDLL with Mfa1 may be stabilized by a charge clamp. These results indicate that the Mfa1-interacting interface of streptococcal antigen I/II encompasses both the KKVQDLLKK and NITVK motif and suggest that the adherence of P. gingivalis to streptococci is driven by a protein-protein interaction domain that resembles the eukaryotic NR box. Thus, both motifs must be taken into account in designing potential peptidomimetics that target P. gingivalis adherence and biofilm formation. .
Figures

References (VSports app下载)
-
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. - V体育官网入口 - PMC - PubMed
-
- Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6383-390. - PubMed
Publication types (VSports)
"VSports在线直播" MeSH terms
- "VSports注册入口" Actions
- VSports在线直播 - Actions
- "V体育官网" Actions
- "VSports在线直播" Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- "V体育2025版" Actions
- Actions (V体育平台登录)
- V体育官网入口 - Actions
"VSports注册入口" Substances
- Actions (V体育ios版)
- V体育官网 - Actions
- "VSports注册入口" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

