VSports注册入口 - Viral IRES RNA structures and ribosome interactions
- PMID: 18468443
- PMCID: PMC2706518
- DOI: 10.1016/j.tibs.2008.04.007
Viral IRES RNA structures and ribosome interactions
Abstract
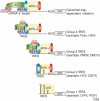
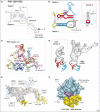
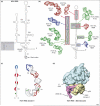
In eukaryotes, protein synthesis initiates primarily by a mechanism that requires a modified nucleotide 'cap' on the mRNA and also proteins that recruit and position the ribosome. Many pathogenic viruses use an alternative, cap-independent mechanism that substitutes RNA structure for the cap and many proteins VSports手机版. The RNAs driving this process are called internal ribosome-entry sites (IRESs) and some are able to bind the ribosome directly using a specific 3D RNA structure. Recent structures of IRES RNAs and IRES-ribosome complexes are revealing the structural basis of viral IRES' 'hijacking' of the protein-making machinery. It now seems that there are fundamental differences in the 3D structures used by different IRESs, although there are some common features in how they interact with ribosomes. .
Figures (VSports)


References
-
- Pestova T, et al. The mechanism of translation initiation in eukaryotes. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 87–128.
-
- Jang SK, et al. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. - VSports手机版 - PMC - PubMed
-
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. - "V体育官网" PubMed
-
- Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 129–153.
-
- Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 155–172.
"VSports app下载" Publication types
MeSH terms
- Actions (VSports app下载)
- "V体育安卓版" Actions
- VSports最新版本 - Actions
Substances
Grants and funding
VSports手机版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

