The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1
- PMID: 18278048
- PMCID: PMC2610020
- DOI: 10.1038/ni1564
The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1
Abstract (V体育安卓版)
Transforming growth factor-beta (TGF-beta) signaling in naive T cells induces expression of the transcription factor Foxp3, a 'master' regulator of regulatory T cells (T(reg) cells). However, the molecular mechanisms leading to Foxp3 induction remain unclear. Here we show that Itch-/- T cells were resistant to TGF-beta treatment and had less Foxp3 expression. The E3 ubiquitin ligase Itch associated with and promoted conjugation of ubiquitin to the transcription factor TIEG1. Itch cooperated with TIEG1 to induce Foxp3 expression, which was reversed by TIEG1 deficiency. Functionally, 'TGF-beta-converted' T(reg) cells generated from TIEG1-deficient mice were unable to suppress airway inflammation in vivo. These results suggest TIEG and Itch contribute to a ubiquitin-dependent nonproteolytic pathway that regulates inducible Foxp3 expression and the control of allergic responses VSports手机版. .
"V体育官网入口" Figures
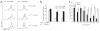






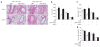
References
-
- Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol. 1999;17:255–281. - PubMed
-
- Umetsu DT, McIntire JJ, Akbari O, Macaubas C, DeKruyff RH. Asthma: an epidemic of dysregulated immunity. Nat Immunol. 2002;3:715–720. - PubMed
-
- Herrick CA, Bottomly K. To respond or not to respond: T cells in allergic asthma. Nat Rev Immunol. 2003;3:405–412. - PubMed
-
- McMenamin C, Pimm C, McKersey M, Holt PG. Regulation of IgE responses to inhaled antigen in mice by antigen-specific γδ T cells. Science. 1994;265:1869–1871. - PubMed
VSports最新版本 - Publication types
"VSports最新版本" MeSH terms
- "V体育ios版" Actions
- "V体育安卓版" Actions
- "VSports注册入口" Actions
- Actions (VSports注册入口)
- "VSports app下载" Actions
- VSports在线直播 - Actions
Substances
- Actions (VSports手机版)
- "VSports app下载" Actions
Grants and funding (V体育安卓版)
LinkOut - more resources
"V体育ios版" Full Text Sources
Other Literature Sources (V体育2025版)
Molecular Biology Databases
Research Materials

