Bcl2 inhibits abasic site repair by down-regulating APE1 endonuclease activity (VSports最新版本)
- PMID: 18263880
- PMCID: PMC2442307
- DOI: 10.1074/jbc.M708345200
Bcl2 inhibits abasic site repair by down-regulating APE1 endonuclease activity
Abstract
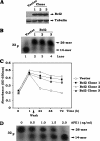
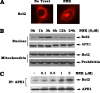
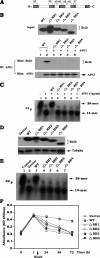
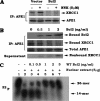
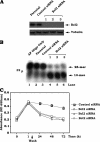
Bcl2 not only prolongs cell survival but also suppresses the repair of abasic (AP) sites of DNA lesions. Apurinic/apyrimidinic endonuclease 1 (APE1) plays a central role in the repair of AP sites via the base excision repair pathway. Here we found that Bcl2 down-regulates APE1 endonuclease activity in association with inhibition of AP site repair. Exposure of cells to nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone results in accumulation of Bcl2 in the nucleus and interaction with APE1, which requires all of the BH domains of Bcl2. Deletion of any of the BH domains from Bcl2 abrogates the ability of Bcl2 to interact with APE1 as well as the inhibitory effects of Bcl2 on APE1 activity and AP site repair. Overexpression of Bcl2 in cells reduces formation of the APE1. XRCC1 complex, and purified Bcl2 protein directly disrupts the APE1. XRCC1 complex with suppression of APE1 endonuclease activity in vitro. Importantly, specific knockdown of endogenous Bcl2 by RNA interference enhances APE1 endonuclease activity with accelerated AP site repair. Thus, Bcl2 inhibition of AP site repair may occur in a novel mechanism by down-regulating APE1 endonuclease activity, which may promote genetic instability and tumorigenesis. VSports手机版.
"VSports" Figures

References (VSports最新版本)
-
- Kelley, M., and Parsons, S. (2001) Antioxid. Redox Signal. 3 671-683 - PubMed
-
- Lau, J., Weatherdon, K., Skalski, V., and Hedley, D. (2004) Br. J. Cancer 91 1166-1173 - PMC (V体育2025版) - PubMed
-
- Tell, G., Damante, G., Caldwell, D., and Kelley, M. (2005) Antioxid. Redox Signal. 7 367-384 - PubMed
-
- Jin, Z., May, W. S., Gao, F., Flagg, T., and Deng, X. (2006) J. Biol. Chem. 281 14446-14456 - PubMed
-
- Xanthoudakis, S., Miao, G., and Curran, T. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 23-27 - PMC (VSports最新版本) - PubMed
"VSports在线直播" Publication types
MeSH terms
- "V体育官网入口" Actions
- Actions (V体育安卓版)
- "VSports" Actions
- Actions (VSports)
- V体育2025版 - Actions
- Actions (VSports注册入口)
- Actions (VSports最新版本)
- Actions (V体育平台登录)
- Actions (V体育ios版)
"VSports在线直播" Substances
- VSports app下载 - Actions
Grants and funding
"V体育官网" LinkOut - more resources
"V体育安卓版" Full Text Sources
Research Materials
Miscellaneous

