"VSports在线直播" Loss of ATRX leads to chromosome cohesion and congression defects
- PMID: 18227278
- PMCID: PMC2213576
- DOI: "V体育官网" 10.1083/jcb.200706083
Loss of ATRX leads to chromosome cohesion and congression defects
Abstract
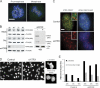
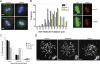
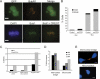
Alpha thalassemia/mental retardation X linked (ATRX) is a switch/sucrose nonfermenting-type ATPase localized at pericentromeric heterochromatin in mouse and human cells. Human ATRX mutations give rise to mental retardation syndromes characterized by developmental delay, facial dysmorphisms, cognitive deficits, and microcephaly and the loss of ATRX in the mouse brain leads to reduced cortical size VSports手机版. We find that ATRX is required for normal mitotic progression in human cultured cells and in neuroprogenitors. Using live cell imaging, we show that the transition from prometaphase to metaphase is prolonged in ATRX-depleted cells and is accompanied by defective sister chromatid cohesion and congression at the metaphase plate. We also demonstrate that loss of ATRX in the embryonic mouse brain induces mitotic defects in neuroprogenitors in vivo with evidence of abnormal chromosome congression and segregation. These findings reveal that ATRX contributes to chromosome dynamics during mitosis and provide a possible cellular explanation for reduced cortical size and abnormal brain development associated with ATRX deficiency. .
Figures


References
-
- Alderton, G.K., L. Galbiati, E. Griffith, K.H. Surinya, H. Neitzel, A.P. Jackson, P.A. Jeggo, and M. O'Driscoll. 2006. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat. Cell Biol. 8:725–733. - PubMed
-
- Baetz, K.K., N.J. Krogan, A. Emili, J. Greenblatt, and P. Hieter. 2004. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24:1232–1244. - "V体育2025版" PMC - PubMed
-
- Bartek, J. 2006. Microcephalin guards against small brains, genetic instability, and cancer. Cancer Cell. 10:91–93. - "VSports在线直播" PubMed
-
- Berube, N.G., C.A. Smeenk, and D.J. Picketts. 2000. Cell cycle-dependant phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet. 9:539–547. - PubMed
Publication types
"V体育ios版" MeSH terms
- V体育2025版 - Actions
- Actions (V体育2025版)
- "V体育官网" Actions
- Actions (V体育ios版)
- Actions (VSports app下载)
- "VSports" Actions
- VSports手机版 - Actions
- "V体育2025版" Actions
- Actions (VSports)
Substances
- "V体育官网入口" Actions
- "V体育平台登录" Actions
LinkOut - more resources (V体育官网入口)
Full Text Sources
Other Literature Sources

