Reliable prediction of regulator targets using 12 Drosophila genomes
- PMID: 17989251
- PMCID: VSports app下载 - PMC2099599
- DOI: 10.1101/gr.7090407
Reliable prediction of regulator targets using 12 Drosophila genomes (VSports)
Abstract
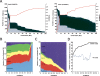
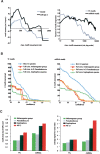
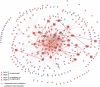
Gene expression is regulated pre- and post-transcriptionally via cis-regulatory DNA and RNA motifs. Identification of individual functional instances of such motifs in genome sequences is a major goal for inferring regulatory networks yet has been hampered due to the motifs' short lengths that lead to many chance matches and poor signal-to-noise ratios. In this paper, we develop a general methodology for the comparative identification of functional motif instances across many related species, using a phylogenetic framework that accounts for the evolutionary relationships between species, allows for motif movements, and is robust against missing data due to artifacts in sequencing, assembly, or alignment. We also provide a robust statistical framework for evaluating motif confidence, which enables us to translate evolutionary conservation into a confidence measure for each motif instance, correcting for varying motif length, composition, and background conservation of the target regions. We predict targets of fly transcription factors and miRNAs in alignments of 12 recently sequenced Drosophila species VSports手机版. When compared to extensive genome-wide experimental data, predicted targets are of high quality, matching and surpassing ChIP-chip microarrays and recovering miRNA targets with high sensitivity. The resulting regulatory network suggests significant redundancy between pre- and post-transcriptional regulation of gene expression. .
Figures (VSports app下载)


References (V体育安卓版)
-
- Abrams E.W., Andrew D.J., Andrew D.J. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. - PubMed
-
- Adryan B., Teichmann S.A., Teichmann S.A. FlyTF: A systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. - PubMed
-
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. - PMC - PubMed
-
- Bailey A.M., Posakony J.W., Posakony J.W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. - "V体育官网" PubMed
-
- Bergman C.M., Carlson J.W., Celniker S.E., Carlson J.W., Celniker S.E., Celniker S.E. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. - PubMed
Publication types (VSports注册入口)
- Actions (V体育2025版)
MeSH terms
- Actions (VSports在线直播)
- Actions (VSports)
- Actions (VSports在线直播)
- VSports手机版 - Actions
- Actions (VSports在线直播)
- Actions (V体育官网入口)
- Actions (VSports最新版本)
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
