Functional genomic delineation of TLR-induced transcriptional networks
- PMID: 17967192
- PMCID: PMC2175519 (V体育安卓版)
- DOI: 10.1186/1471-2164-8-394
Functional genomic delineation of TLR-induced transcriptional networks
Abstract
Background: The innate immune system is the first line of defense mechanisms protecting the host from invading pathogens such as bacteria and viruses. The innate immunity responses are triggered by recognition of prototypical pathogen components by cellular receptors. Prominent among these pathogen sensors are Toll-like receptors (TLRs) VSports手机版. We sought global delineation of transcriptional networks induced by TLRs, analyzing four genome-wide expression datasets in mouse and human macrophages stimulated with pathogen-mimetic agents that engage various TLRs. .
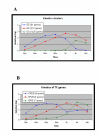
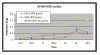
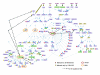
Results: Combining computational analysis of expression profiles and cis-regulatory promoter sequences, we dissected the TLR-induced transcriptional program into two major components: the first is universally activated by all examined TLRs, and the second is specific to activated TLR3 and TLR4. Our results point to NF-kappaB and ISRE-binding transcription factors as the key regulators of the universal and the TLR3/4-specific responses, respectively, and identify novel putative positive and negative feedback loops in these transcriptional programs V体育安卓版. Analysis of the kinetics of the induced network showed that while NF-kappaB regulates mainly an early-induced and sustained response, the ISRE element functions primarily in the induction of a delayed wave. We further demonstrate that co-occurrence of the NF-kappaB and ISRE elements in the same promoter endows its targets with enhanced responsiveness. .
Conclusion: Our results enhance system-level understanding of the networks induced by TLRs and demonstrate the power of genomics approaches to delineate intricate transcriptional webs in mammalian systems. Such systems-level knowledge of the TLR network can be useful for designing ways to pharmacologically manipulate the activity of the innate immunity in pathological conditions in which either enhancement or repression of this branch of the immune system is desired. V体育ios版.
Figures


References
-
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/S1074-7613(02)00390-4. - V体育安卓版 - DOI - PubMed
-
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. - DOI - PMC - PubMed
Publication types (VSports手机版)
- "V体育官网" Actions
MeSH terms
- V体育安卓版 - Actions
- "VSports app下载" Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
Substances
- Actions (V体育安卓版)
LinkOut - more resources (VSports注册入口)
Full Text Sources

