Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis
- PMID: 17889646
- PMCID: PMC2586609
- DOI: 10.1016/j.cell.2007.07.020
Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis
"V体育ios版" Abstract
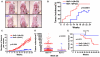
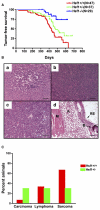
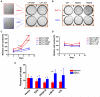
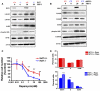
Heat shock factor 1 (HSF1) is the master regulator of the heat shock response in eukaryotes, a very highly conserved protective mechanism. HSF1 function increases survival under a great many pathophysiological conditions. How it might be involved in malignancy remains largely unexplored. We report that eliminating HSF1 protects mice from tumors induced by mutations of the RAS oncogene or a hot spot mutation in the tumor suppressor p53. In cell culture, HSF1 supports malignant transformation by orchestrating a network of core cellular functions including proliferation, survival, protein synthesis, and glucose metabolism. The striking effects of HSF1 on oncogenic transformation are not limited to mouse systems or tumor initiation; human cancer lines of diverse origins show much greater dependence on HSF1 function to maintain proliferation and survival than their nontransformed counterparts. While it enhances organismal survival and longevity under most circumstances, HSF1 has the opposite effect in supporting the lethal phenomenon of cancer. VSports手机版.
Figures


Comment in
-
Non-oncogene addiction and the stress phenotype of cancer cells.Cell. 2007 Sep 21;130(6):986-8. doi: 10.1016/j.cell.2007.09.007. Cell. 2007. PMID: 17889643
References
-
- Auluck PK, Meulener MC, Bonini N. Mechanisms of suppression of {alpha}-synuclein neurotoxicity by geldanamycin in Drosophila. J. Biol. Chem. 2005;280:2873–2878. - "VSports最新版本" PubMed
-
- Balmain A, Ramsden M, Bowden GT, Smith J. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–660. - PubMed (V体育平台登录)
-
- Beausejour CM, Campisi J. Ageing: balancing regeneration and cancer. Nature. 2006;443:404–405. - PubMed
-
- Bissell MJ, Rambeck WA, White RC, Bassham JA. Glycerol phosphate shuttle in virus-transformed cells in culture. Science. 1976;191:856–858. - VSports最新版本 - PubMed
Publication types
- Actions (V体育ios版)
MeSH terms
- "V体育2025版" Actions
- "V体育官网" Actions
- V体育平台登录 - Actions
- VSports最新版本 - Actions
- "V体育官网入口" Actions
- Actions (VSports在线直播)
- "V体育安卓版" Actions
- VSports注册入口 - Actions
- Actions (V体育2025版)
- "VSports app下载" Actions
- V体育平台登录 - Actions
- V体育平台登录 - Actions
- "VSports注册入口" Actions
- "V体育官网" Actions
- "VSports app下载" Actions
- Actions (V体育平台登录)
- "VSports app下载" Actions
- V体育官网入口 - Actions
- VSports app下载 - Actions
- VSports app下载 - Actions
- "V体育官网" Actions
Substances
- "VSports最新版本" Actions
- "VSports在线直播" Actions
- VSports注册入口 - Actions
- Actions (V体育官网)
- "VSports最新版本" Actions
- VSports - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources (VSports在线直播)
Medical
"VSports" Molecular Biology Databases
Research Materials (V体育官网入口)
Miscellaneous (VSports app下载)

