Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes
- PMID: 17452440
- PMCID: PMC1951502
- DOI: 10.1128/MCB.02432-06
Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes
Abstract
DET1 (de-etiolated 1) is an essential negative regulator of plant light responses, and it is a component of the Arabidopsis thaliana CDD complex containing DDB1 and COP10 ubiquitin E2 variant. Human DET1 has recently been isolated as one of the DDB1- and Cul4A-associated factors, along with an array of WD40-containing substrate receptors of the Cul4A-DDB1 ubiquitin ligase. However, DET1 differs from conventional substrate receptors of cullin E3 ligases in both biochemical behavior and activity. Here we report that mammalian DET1 forms stable DDD-E2 complexes, consisting of DDB1, DDA1 (DET1, DDB1 associated 1), and a member of the UBE2E group of canonical ubiquitin-conjugating enzymes. DDD-E2 complexes interact with multiple ubiquitin E3 ligases. We show that the E2 component cannot maintain the ubiquitin thioester linkage once bound to the DDD core, rendering mammalian DDD-E2 equivalent to the Arabidopsis CDD complex. While free UBE2E-3 is active and able to enhance UbcH5/Cul4A activity, the DDD core specifically inhibits Cul4A-dependent polyubiquitin chain assembly in vitro. Overexpression of DET1 inhibits UV-induced CDT1 degradation in cultured cells VSports手机版. These findings demonstrate that the conserved DET1 complex modulates Cul4A functions by a novel mechanism. .
Figures

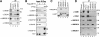



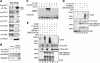


V体育2025版 - References
-
- Angers, S., T. Li, X. Yi, M. J. Maccoss, R. T. Moon, and N. Zheng. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443:590-593. - PubMed
-
- Benvenuto, G., F. Formiggini, P. Laflamme, M. Malakhov, and C. Bowler. 2002. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 12:1529-1534. - PubMed (V体育官网)
-
- Bernhardt, A., E. Lechner, P. Hano, V. Schade, M. Dieterle, M. Anders, M. J. Dubin, G. Benvenuto, C. Bowler, P. Genschik, and H. Hellmann. 2006. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47:591-603. - PubMed
-
- Bondar, T., E. V. Mirkin, D. S. Ucker, W. E. Walden, S. M. Mirkin, and P. Raychaudhuri. 2003. Schizosaccharomyces pombe Ddb1 is functionally linked to the replication checkpoint pathway. J. Biol. Chem. 278:37006-37014. - PubMed (VSports手机版)
Publication types
- Actions (VSports在线直播)
V体育安卓版 - MeSH terms
- "V体育官网入口" Actions
- "VSports" Actions
- V体育2025版 - Actions
- V体育ios版 - Actions
- VSports app下载 - Actions
- Actions (VSports)
- VSports在线直播 - Actions
- Actions (V体育官网)
- "VSports注册入口" Actions
- Actions (V体育官网)
- "V体育ios版" Actions
Substances (V体育2025版)
- Actions (VSports注册入口)
- VSports app下载 - Actions
- VSports - Actions
- "VSports在线直播" Actions
- "V体育官网" Actions
- "V体育安卓版" Actions
"V体育ios版" Grants and funding
LinkOut - more resources
Full Text Sources
VSports注册入口 - Other Literature Sources
Molecular Biology Databases (VSports app下载)
Research Materials
