Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins
- PMID: 17437998
- PMCID: PMC1847712
- DOI: "VSports" 10.1101/gad.1528507
V体育2025版 - Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins
Abstract
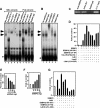
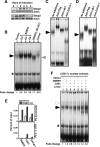
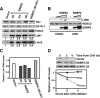
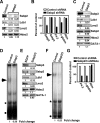
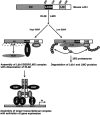
The LIM domain-binding protein Ldb1 is an essential cofactor of LIM-homeodomain (LIM-HD) and LIM-only (LMO) proteins in development. The stoichiometry of Ldb1, LIM-HD, and LMO proteins is tightly controlled in the cell and is likely a critical determinant of their biological actions. Single-stranded DNA-binding proteins (SSBPs) were recently shown to interact with Ldb1 and are also important in developmental programs. We establish here that two mammalian SSBPs, SSBP2 and SSBP3, contribute to an erythroid DNA-binding complex that contains the transcription factors Tal1 and GATA-1, the LIM domain protein Lmo2, and Ldb1 and binds a bipartite E-box-GATA DNA sequence motif. In addition, SSBP2 was found to augment transcription of the Protein 4. 2 (P4. 2) gene, a direct target of the E-box-GATA-binding complex, in an Ldb1-dependent manner and to increase endogenous Ldb1 and Lmo2 protein levels, E-box-GATA DNA-binding activity, and P4. 2 and beta-globin expression in erythroid progenitors. Finally, SSBP2 was demonstrated to inhibit Ldb1 and Lmo2 interaction with the E3 ubiquitin ligase RLIM, prevent RLIM-mediated Ldb1 ubiquitination, and protect Ldb1 and Lmo2 from proteasomal degradation. These results define a novel biochemical function for SSBPs in regulating the abundance of LIM domain and LIM domain-binding proteins VSports手机版. .
Figures

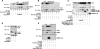
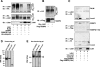
References
-
- Agulnick A.D., Taira M., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Taira M., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Breen J.J., Tanaka T., Dawid I.B., Westphal H., Tanaka T., Dawid I.B., Westphal H., Dawid I.B., Westphal H., Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. - VSports最新版本 - PubMed
-
- Amerik A.Y., Hochstrasser M., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. - PubMed
-
- Aoyama M., Ozaki T., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Ozaki T., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Inuzuka H., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Tomotsune D., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Hirato J., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Okamoto Y., Tokita H., Ohira M., Nakagawara A., Tokita H., Ohira M., Nakagawara A., Ohira M., Nakagawara A., Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. - PubMed (VSports手机版)
-
- Bach I., Carriere C., Ostendorff H.P., Andersen B., Rosenfeld M.G., Carriere C., Ostendorff H.P., Andersen B., Rosenfeld M.G., Ostendorff H.P., Andersen B., Rosenfeld M.G., Andersen B., Rosenfeld M.G., Rosenfeld M.G. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes & Dev. 1997;11:1370–1380. - PubMed
Publication types
- Actions (V体育2025版)
- "VSports手机版" Actions
MeSH terms
- "VSports app下载" Actions
- "VSports最新版本" Actions
- Actions (VSports app下载)
- VSports注册入口 - Actions
- Actions (VSports在线直播)
- VSports手机版 - Actions
- Actions (V体育2025版)
- VSports - Actions
- "VSports注册入口" Actions
- VSports在线直播 - Actions
- VSports在线直播 - Actions
Substances
- "V体育2025版" Actions
- VSports - Actions
- V体育官网入口 - Actions
- "VSports在线直播" Actions
Grants and funding
"VSports手机版" LinkOut - more resources
V体育安卓版 - Full Text Sources
