V体育平台登录 - QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli
- PMID: 17339361
- PMCID: V体育2025版 - PMC1865749
- DOI: 10.1128/IAI.02003-06 (V体育安卓版)
QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli
Abstract
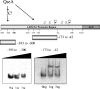
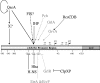
Quorum sensing (QS) in enterohemorrhagic Escherichia coli (EHEC) regulates the expression of the locus of enterocyte effacement (LEE). The LEE contains five major operons named LEE1 through LEE5. QseA was previously shown to be activated through QS and to activate the transcription of LEE1. The LEE1 operon encodes Ler, the transcription activator of all other LEE genes, and has two promoters: a distal promoter (P1) and a proximal promoter (P2). We have previously reported that QseA acts on P1 and not P2. To identify the minimal region of LEE1 that is necessary for QseA-mediated activation, a series of nested-deletion constructs of the LEE1 promoter fused to a lacZ reporter were constructed in both the EHEC and E. coli K-12 backgrounds. In an EHEC background, QseA-dependent activation of LEE1 can be observed for the entire regulatory region (beginning at nucleotide -393 and ending at nucleotide -123). In contrast to what occurred in EHEC, in K-12 there was no QseA-dependent activation of LEE1 transcription between base pairs -393 and -343 VSports手机版. These data indicate that a QseA-dependent EHEC-specific regulator is required for the activation of transcription in this region. We also observed QseA-dependent LEE1 activation from nucleotides -218 to -123 in K-12, similar to results of the nested-deletion analysis performed with EHEC. Electrophoretic mobility shift assays established that QseA directly binds to the region of LEE1 from bp -173 to -42 and not to the region from bp -393 to -343. These studies suggest that QseA activates the transcription of LEE1 by directly binding upstream of its P1 promoter region. .
Figures




References
-
- Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. - "V体育平台登录" PubMed
-
- Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:217-228. - PubMed
Publication types
- "V体育官网入口" Actions
- VSports注册入口 - Actions
MeSH terms
- VSports在线直播 - Actions
- VSports最新版本 - Actions
- Actions (VSports)
- "V体育官网入口" Actions
- Actions (VSports在线直播)
- "VSports app下载" Actions
- VSports注册入口 - Actions
- "V体育安卓版" Actions
- V体育平台登录 - Actions
Substances
- VSports app下载 - Actions
- V体育安卓版 - Actions
Grants and funding (V体育官网)
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育平台登录)

