Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities
- PMID: 17220268
- PMCID: "V体育安卓版" PMC1828774
- DOI: "VSports app下载" 10.1128/AEM.01996-06
"VSports最新版本" Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities
Abstract
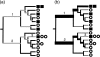
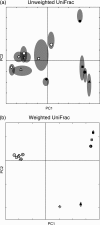
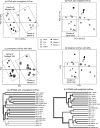
The assessment of microbial diversity and distribution is a major concern in environmental microbiology. There are two general approaches for measuring community diversity: quantitative measures, which use the abundance of each taxon, and qualitative measures, which use only the presence/absence of data. Quantitative measures are ideally suited to revealing community differences that are due to changes in relative taxon abundance (e. g. , when a particular set of taxa flourish because a limiting nutrient source becomes abundant). Qualitative measures are most informative when communities differ primarily by what can live in them (e. g. , at high temperatures), in part because abundance information can obscure significant patterns of variation in which taxa are present. We illustrate these principles using two 16S rRNA-based surveys of microbial populations and two phylogenetic measures of community beta diversity: unweighted UniFrac, a qualitative measure, and weighted UniFrac, a new quantitative measure, which we have added to the UniFrac website (http://bmf. colorado. edu/unifrac). These studies considered the relative influences of mineral chemistry, temperature, and geography on microbial community composition in acidic thermal springs in Yellowstone National Park and the influences of obesity and kinship on microbial community composition in the mouse gut. We show that applying qualitative and quantitative measures to the same data set can lead to dramatically different conclusions about the main factors that structure microbial diversity and can provide insight into the nature of community differences. We also demonstrate that both weighted and unweighted UniFrac measurements are robust to the methods used to build the underlying phylogeny VSports手机版. .
Figures



References
-
- Badano, E. I., and L. A. Cavieres. 2006. Impacts of ecosystem engineers on community attributes: effects of cushion plants at different elevations of the Chilean Andes. Divers. Distrib. 12:388-396.
-
- Bluis, J., and D. Shin. 2003. Nodal distance algorithm: calculating a phylogenetic tree comparison metric, p. 87-94. In Proceedings of the Third IEEE Symposium on BioInformatics and BioEngineering. IEEE, Los Alamitos, CA.
-
- De Benedictis, P. A. 1973. On the correlations between certain diversity indices. Am. Nat. 107:295-302.
-
- Felsenstein, J. 2004. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, MA.
Publication types (V体育安卓版)
- "V体育ios版" Actions
MeSH terms
- Actions (V体育平台登录)
- Actions (VSports)
- "V体育官网" Actions
- Actions (V体育平台登录)
- V体育2025版 - Actions
- Actions (VSports注册入口)
- Actions (V体育安卓版)
- V体育2025版 - Actions
- "V体育安卓版" Actions
Substances
- VSports注册入口 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources

