"V体育官网入口" Human cancers express a mutator phenotype
- PMID: 17108085
- PMCID: PMC1636340
- DOI: "VSports最新版本" 10.1073/pnas.0607057103
"VSports app下载" Human cancers express a mutator phenotype
Erratum in
- Proc Natl Acad Sci U S A. 2006 Dec 19;103(51):19605
Abstract
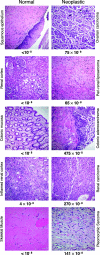
Cancer cells contain numerous clonal mutations, i. e. , mutations that are present in most or all malignant cells of a tumor and have presumably been selected because they confer a proliferative advantage. An important question is whether cancer cells also contain a large number of random mutations, i. e. , randomly distributed unselected mutations that occur in only one or a few cells of a tumor. Such random mutations could contribute to the morphologic and functional heterogeneity of cancers and include mutations that confer resistance to therapy. We have postulated that malignant cells exhibit a mutator phenotype resulting in the generation of random mutations throughout the genome. We have recently developed an assay to quantify random mutations in human tissue with unprecedented sensitivity. Here, we report measurements of random single-nucleotide substitutions in normal and neoplastic human tissues. In normal tissues, the frequency of spontaneous random mutations is exceedingly low, less than 1 x 10(-8) per base pair VSports手机版. In contrast, tumors from the same individuals exhibited an average frequency of 210 x 10(-8) per base pair, an elevation of at least two orders of magnitude. Our data document tumor heterogeneity at the single-nucleotide level, indicate that accelerated mutagenesis prevails late into tumor progression, and suggest that elevation of random mutation frequency in tumors might serve as a novel prognostic indicator. .
"VSports app下载" Conflict of interest statement
The authors declare no conflict of interest.
"V体育官网" Figures


Comment in
-
Random mutations, selected mutations: A PIN opens the door to new genetic landscapes.Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18033-4. doi: 10.1073/pnas.0609000103. Epub 2006 Nov 20. Proc Natl Acad Sci U S A. 2006. PMID: 17116890 Free PMC article. No abstract available.
References
-
- Weinberg RA. Biology of Cancer. New York: Garland Science; 2007.
-
- Levine AJ. Cell. 1997;88:323–331. - PubMed
-
- Hussain SP, Amstad P, Raja K, Ambs S, Nagashima M, Bennett WP, Shields PG, Ham A-J, Swenberg JA, Marrogi AJ, Harris CC. Cancer Res. 2000;60:3333–3337. - PubMed
-
- Barbacid M. Annu Rev Biochem. 1987;56:779–827. - PubMed
-
- Albertson DG, Collins C, McCormick F, Gray JW. Nat Genet. 2003;34:369–376. - PubMed
Publication types (V体育2025版)
MeSH terms
- VSports在线直播 - Actions
- V体育ios版 - Actions
- "V体育ios版" Actions
- Actions (VSports)
Substances
Grants and funding
"V体育ios版" LinkOut - more resources
Full Text Sources
VSports最新版本 - Other Literature Sources

