"V体育官网" Conservation and evolution of gene coexpression networks in human and chimpanzee brains
- PMID: 17101986
- PMCID: "V体育官网入口" PMC1693857
- DOI: 10.1073/pnas.0605938103 (VSports)
Conservation and evolution of gene coexpression networks in human and chimpanzee brains (V体育安卓版)
Abstract
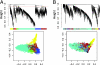
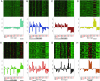
Comparisons of gene expression between human and non-human primate brains have identified hundreds of differentially expressed genes, yet translating these lists into key functional distinctions between species has proved difficult. Here we provide a more integrated view of human brain evolution by examining the large-scale organization of gene coexpression networks in human and chimpanzee brains. We identify modules of coexpressed genes that correspond to discrete brain regions and quantify their conservation between the species VSports手机版. Module conservation in cerebral cortex is significantly weaker than module conservation in subcortical brain regions, revealing a striking gradient that parallels known evolutionary hierarchies. We introduce a method for identifying species-specific network connections and demonstrate how differential network connectivity can be used to identify key drivers of evolutionary change. By integrating our results with comparative genomic sequence data and estimates of protein sequence divergence rates, we confirm a number of network predictions and validate these findings. Our results provide insights into the molecular bases of primate brain organization and demonstrate the general utility of weighted gene coexpression network analysis. .
Conflict of interest statement
The authors declare no conflict of interest.
Figures
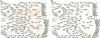

References
-
- Deacon TW. The Symbolic Species: The Co-evolution of Language and the Brain. New York, NY: Norton; 1997.
-
- King MC, Wilson AC. Science. 1975;188:107–116. - PubMed
-
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Science. 2002;296:340–343. - PubMed
"V体育2025版" Publication types
"VSports最新版本" MeSH terms
- VSports手机版 - Actions
- "V体育平台登录" Actions
- "VSports app下载" Actions
- V体育安卓版 - Actions
- "VSports app下载" Actions
- "VSports最新版本" Actions
Grants and funding (VSports注册入口)
LinkOut - more resources
Full Text Sources
"VSports手机版" Other Literature Sources
V体育安卓版 - Molecular Biology Databases

