V体育官网入口 - Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient
- PMID: 16707682
- PMCID: PMC1482900
- DOI: 10.1128/JB.01974-05
"VSports最新版本" Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient
Abstract
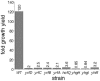
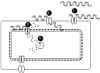
Natural genetic competence is the ability of cells to take up extracellular DNA and is an important mechanism for horizontal gene transfer. Another potential benefit of natural competence is that exogenous DNA can serve as a nutrient source for starving bacteria because the ability to "eat" DNA is necessary for competitive survival in environments containing limited nutrients. We show here that eight Escherichia coli genes, identified as homologs of com genes in Haemophilus influenzae and Neisseria gonorrhoeae, are necessary for the use of extracellular DNA as the sole source of carbon and energy VSports手机版. These genes also confer a competitive advantage to E. coli during long-term stationary-phase incubation. We also show that homologs of these genes are found throughout the proteobacteria, suggesting that the use of DNA as a nutrient may be a widespread phenomenon. .
Figures
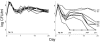

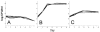
References
-
- Averhoff, B., and A. Friedrich. 2003. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180:385-393. - PubMed
-
- Bauer, F., C. Hertel, and W. P. Hammes. 1999. Transformation of Escherichia coli in foodstuffs. Syst. Appl. Microbiol. 22:161-168. - PubMed
-
- Beacham, I. R., E. Yagil, K. Beacham, and R. H. Pritchard. 1971. On the localization of enzymes of deoxynucleoside catabolism in Escherichia coli. Mol. Gen. Genet. 102:112-127. - PubMed
-
- Benz, R., A. Schmid, C. Maier, and E. Bremer. 1988. Characterization of the nucleoside-binding site inside the Tsx channel of Escherichia coli outer membrane. Reconstitution experiments with lipid bilayer membranes. Eur. J. Biochem. 176:699-705. - PubMed
Publication types
MeSH terms
- "V体育ios版" Actions
- "V体育官网" Actions
- Actions (VSports手机版)
- VSports手机版 - Actions
- "V体育官网入口" Actions
"V体育官网" Substances
- "VSports注册入口" Actions
V体育2025版 - Grants and funding
LinkOut - more resources (VSports)
VSports手机版 - Full Text Sources
Other Literature Sources
Molecular Biology Databases

