Cell-type specific gene expression profiles of leukocytes in human peripheral blood
- PMID: 16704732
- PMCID: PMC1479811
- DOI: "V体育2025版" 10.1186/1471-2164-7-115
Cell-type specific gene expression profiles of leukocytes in human peripheral blood (VSports注册入口)
"V体育官网入口" Abstract
Background: Blood is a complex tissue comprising numerous cell types with distinct functions and corresponding gene expression profiles VSports手机版. We attempted to define the cell type specific gene expression patterns for the major constituent cells of blood, including B-cells, CD4+ T-cells, CD8+ T-cells, lymphocytes and granulocytes. We did this by comparing the global gene expression profiles of purified B-cells, CD4+ T-cells, CD8+ T-cells, granulocytes, and lymphocytes using cDNA microarrays. .
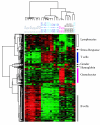
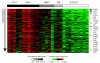
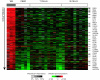
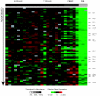
Results: Unsupervised clustering analysis showed that similar cell populations from different donors share common gene expression profiles. Supervised analyses identified gene expression signatures for B-cells (427 genes), T-cells (222 genes), CD8+ T-cells (23 genes), granulocytes (411 genes), and lymphocytes (67 genes). No statistically significant gene expression signature was identified for CD4+ cells. Genes encoding cell surface proteins were disproportionately represented among the genes that distinguished among the lymphocyte subpopulations. Lymphocytes were distinguishable from granulocytes based on their higher levels of expression of genes encoding ribosomal proteins, while granulocytes exhibited characteristic expression of various cell surface and inflammatory proteins. V体育安卓版.
Conclusion: The genes comprising the cell-type specific signatures encompassed many of the genes already known to be involved in cell-type specific processes, and provided clues that may prove useful in discovering the functions of many still unannotated genes V体育ios版. The most prominent feature of the cell type signature genes was the enrichment of genes encoding cell surface proteins, perhaps reflecting the importance of specialized systems for sensing the environment to the physiology of resting leukocytes. .
Figures


References
-
- Parks D, Herzenberg L. Fluorescence-activated cell sorting: Theory, experimental optimization, and applications in lymphoid cell biology. Methods Enzymol. 1984;108:197–241. - PubMed
-
- Bomprezzi R, Ringner M, Kim S, Bittner ML, Khan J, Chen Y, Elkahloun A, Yu A, Bielekova B, Meltzer PS, Martin R, McFarland HF, Trent JM. Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum Mol Genet. 2003;12:2191–2199. doi: 10.1093/hmg/ddg221. - DOI - PubMed
-
- Connolly PH, Caiozzo VJ, Zaldivar F, Nemet D, Larson J, Hung SP, Heck JD, Hatfield GW, Cooper DM. Effects of exercise on gene expression in human peripheral blood mononuclear cells. J Appl Physiol. 2004;97:1461–1469. doi: 10.1152/japplphysiol.00316.2004. - "VSports最新版本" DOI - PubMed
-
- Lampe JW, Stepaniants SB, Mao M, Radich JP, Dai H, Linsley PS, Friend SH, Potter JD. Signatures of environmental exposures using peripheral leukocyte gene expression: tobacco smoke. Cancer Epidemiol Biomarkers Prev. 2004;13:445–453. - VSports在线直播 - PubMed
-
- Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr, Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu H, Goldin E, Baird AE. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. - DOI - PubMed
Publication types
- VSports在线直播 - Actions
MeSH terms
- Actions (VSports app下载)
- Actions (VSports手机版)
- "V体育ios版" Actions
- Actions (V体育2025版)
- "V体育安卓版" Actions
- "V体育2025版" Actions
- Actions (V体育官网入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
V体育2025版 - Molecular Biology Databases
Research Materials

