The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation (V体育官网入口)
- PMID: 16314513
- PMCID: PMC1316965 (VSports注册入口)
- DOI: "V体育ios版" 10.1128/MCB.25.24.10895-10906.2005
The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation
"VSports app下载" Abstract
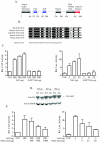
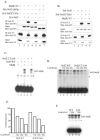
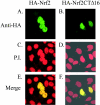
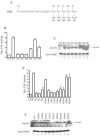
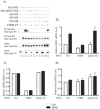
Nrf2 is a transcription factor critical for the maintenance of cellular redox homeostasis. We have previously found that Nrf2 is a labile protein, and its activation in cells under stress involves mechanisms leading to its stabilization. As a modular protein, Nrf2 possesses distinct transactivation and DNA binding domains essential for its transcriptional activity. In this study, we found that the C-terminal "Neh3" domain of Nrf2 is also important for its activity. Deletion of the last 16 amino acids of the protein completely abolishes its ability to activate both reporter and endogenous gene expression. Using site-directed mutagenesis, we have identified a stretch of amino acids within this region that are essential for its activity and that are found to be conserved across species and among other members of the CNC-bZIP family. Importantly, deletion of the final 16 amino acids of Nrf2 does not influence its dimerizing capability, DNA binding activity, or subcellular localization, although it does increase the half-life of the protein. In addition, this region was found to be important for interaction with CHD6 (a chromo-ATPase/helicase DNA binding protein) in a yeast two-hybrid screen VSports手机版. RNA interference-mediated knockdown of CHD6 reduced both the basal and tert-butylhydroquinone-inducible expression of NQO1, a prototypical Nrf2 target gene. These data suggest that the Neh3 domain may act as a transactivation domain and that it is possibly involved in interaction with components of the transcriptional apparatus to affect its transcriptional activity. .
V体育官网入口 - Figures



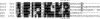
References
-
- Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Capcn'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. - PubMed
-
- Cho, H. Y., A. E. Jedlicka, S. P. Reddy, T. W. Kensler, M. Yamamoto, L. Y. Zhang, and S. R. Kleeberger. 2002. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell. Mol. Biol. 26:175-182. - PubMed
-
- Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. - PubMed
MeSH terms
- V体育2025版 - Actions
- Actions (VSports app下载)
- "V体育官网入口" Actions
- VSports在线直播 - Actions
- "VSports手机版" Actions
- Actions (VSports注册入口)
- VSports - Actions
"VSports最新版本" Substances
- Actions (VSports手机版)
- Actions (V体育2025版)
- Actions (V体育ios版)
- Actions (VSports在线直播)
LinkOut - more resources (V体育官网)
Full Text Sources (V体育官网)
Other Literature Sources
V体育安卓版 - Molecular Biology Databases
Research Materials
Miscellaneous
