Swimming in circles: motion of bacteria near solid boundaries
- PMID: 16239332
- PMCID: PMC1367047 (VSports注册入口)
- DOI: "VSports注册入口" 10.1529/biophysj.105.069401
Swimming in circles: motion of bacteria near solid boundaries (V体育官网)
Abstract
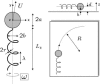
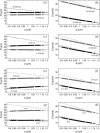
Near a solid boundary, Escherichia coli swims in clockwise circular motion VSports手机版. We provide a hydrodynamic model for this behavior. We show that circular trajectories are natural consequences of force-free and torque-free swimming and the hydrodynamic interactions with the boundary, which also leads to a hydrodynamic trapping of the cells close to the surface. We compare the results of the model with experimental data and obtain reasonable agreement. In particular, the radius of curvature of the trajectory is observed to increase with the length of the bacterium body. .
Figures
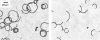




References
-
- Berg, H. C. 2000. Motile behavior of bacteria. Phys. Today. 53:24–29.
-
- Berg, H. C. 2004. Escherichia coli in Motion. Springer-Verlag, New York.
-
- Berg, H. C., and L. Turner. 1990. Chemotaxis of bacteria in glass capillary arrays—Escherichia coli, motility, microchannel plate, and light scattering. Biophys. J. 58:919–930. - "V体育ios版" PMC - PubMed
Publication types
- Actions (VSports在线直播)
- Actions (V体育平台登录)
MeSH terms
- Actions (VSports最新版本)
- VSports app下载 - Actions
- Actions (V体育安卓版)
- V体育2025版 - Actions
- VSports app下载 - Actions
- VSports最新版本 - Actions
- Actions (VSports手机版)
- Actions (V体育安卓版)
- "V体育2025版" Actions
- Actions (VSports)
- "VSports在线直播" Actions
- Actions (VSports)
- "V体育安卓版" Actions
Grants and funding (VSports最新版本)
LinkOut - more resources
Full Text Sources
Other Literature Sources

