Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia
- PMID: 16085880
- PMCID: PMC1183352
- DOI: 10.1128/AEM.71.8.4822-4832.2005
Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia
Abstract
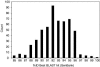
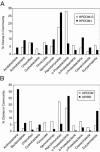
Stromatolites, organosedimentary structures formed by microbial activity, are found throughout the geological record and are important markers of biological history. More conspicuous in the past, stromatolites occur today in a few shallow marine environments, including Hamelin Pool in Shark Bay, Western Australia. Hamelin Pool stromatolites often have been considered contemporary analogs to ancient stromatolites, yet little is known about the microbial communities that build them. We used DNA-based molecular phylogenetic methods that do not require cultivation to study the microbial diversity of an irregular stromatolite and of the surface and interior of a domal stromatolite. To identify the constituents of the stromatolite communities, small subunit rRNA genes were amplified by PCR from community genomic DNA with universal primers, cloned, sequenced, and compared to known rRNA genes. The communities were highly diverse and novel. The average sequence identity of Hamelin Pool sequences compared to the >200,000 known rRNA sequences was only approximately 92% VSports手机版. Clone libraries were approximately 90% bacterial and approximately 10% archaeal, and eucaryotic rRNA genes were not detected in the libraries. The most abundant sequences were representative of novel proteobacteria (approximately 28%), planctomycetes ( approximately 17%), and actinobacteria (approximately 14%). Sequences representative of cyanobacteria, long considered to dominate these communities, comprised <5% of clones. Approximately 10% of the sequences were most closely related to those of alpha-proteobacterial anoxygenic phototrophs. These results provide a framework for understanding the kinds of organisms that build contemporary stromatolites, their ecology, and their relevance to stromatolites preserved in the geological record. .
"V体育2025版" Figures





References
-
- Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. - PubMed
-
- Al-Qassab, S., W. J. Lee, S. Murray, A. G. B. Simpson, and D. J. Patterson. 2002. Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozool. 41:91-144.
Publication types
- "V体育ios版" Actions
- Actions (V体育平台登录)
"VSports app下载" MeSH terms
- "VSports手机版" Actions
- Actions (V体育平台登录)
- "VSports" Actions
- Actions (VSports注册入口)
- Actions (VSports在线直播)
- Actions (V体育官网)
- Actions (V体育官网入口)
- V体育官网 - Actions
- VSports app下载 - Actions
- VSports app下载 - Actions
Substances
"VSports最新版本" Associated data
- Actions
- Actions (V体育安卓版)
- "VSports" Actions
- "V体育官网入口" Actions
- Actions
- VSports注册入口 - Actions
- Actions
- Actions
- Actions (V体育安卓版)
- "V体育安卓版" Actions
- Actions
- Actions (VSports手机版)
- Actions
- "VSports手机版" Actions
- Actions
- Actions
- Actions (VSports app下载)
- "V体育官网" Actions
- VSports - Actions
- Actions
- Actions (V体育官网入口)
- Actions
- VSports app下载 - Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
- VSports - Actions
- Actions (VSports注册入口)
- Actions (VSports手机版)
- Actions
- "VSports app下载" Actions
- VSports在线直播 - Actions
- "VSports app下载" Actions
- Actions
- "VSports" Actions
- VSports app下载 - Actions
- Actions (V体育2025版)
- "V体育ios版" Actions
- "V体育官网入口" Actions
- Actions
- "VSports手机版" Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- Actions
- Actions
- V体育官网入口 - Actions
- Actions (V体育ios版)
- V体育平台登录 - Actions
- V体育官网 - Actions
- VSports - Actions
- "V体育2025版" Actions
- "V体育ios版" Actions
- Actions
- VSports app下载 - Actions
- VSports - Actions
- "V体育安卓版" Actions
- "V体育ios版" Actions
- V体育ios版 - Actions
- Actions
- V体育官网 - Actions
- Actions
- "V体育ios版" Actions
- Actions
- "V体育ios版" Actions
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- Actions
- Actions
- VSports注册入口 - Actions
- Actions (V体育2025版)
- Actions
- Actions
- "V体育2025版" Actions
- VSports注册入口 - Actions
- Actions
- Actions (VSports在线直播)
- "V体育ios版" Actions
- "VSports手机版" Actions
- Actions
- Actions
- Actions
- "V体育平台登录" Actions
- Actions (VSports)
- Actions
- Actions (V体育官网入口)
- VSports - Actions
- Actions (V体育官网入口)
- "V体育官网入口" Actions
- Actions
- Actions
- Actions
- "V体育安卓版" Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- "VSports手机版" Actions
- Actions
- V体育安卓版 - Actions
- Actions
- Actions
- VSports在线直播 - Actions
- "VSports在线直播" Actions
- VSports手机版 - Actions
- Actions
- Actions
- Actions
- "V体育官网入口" Actions
- "V体育2025版" Actions
- Actions
- Actions
- "VSports" Actions
- Actions
- Actions (VSports注册入口)
- "VSports" Actions
- Actions
- "V体育平台登录" Actions
- Actions (V体育平台登录)
- "VSports手机版" Actions
- Actions (V体育安卓版)
- Actions (VSports在线直播)
- Actions
- "V体育ios版" Actions
- Actions
- V体育官网 - Actions
- Actions (V体育ios版)
- Actions
- Actions (V体育官网)
- Actions
- Actions (VSports最新版本)
- Actions (V体育官网)
- Actions
- VSports注册入口 - Actions
- Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- Actions
- VSports手机版 - Actions
- "VSports在线直播" Actions
- "VSports最新版本" Actions
- Actions (VSports app下载)
- V体育ios版 - Actions
- Actions
- VSports最新版本 - Actions
- "VSports" Actions
- Actions
- Actions (VSports最新版本)
- Actions (V体育官网)
- Actions (VSports最新版本)
- Actions
- VSports在线直播 - Actions
- Actions (VSports app下载)
- Actions
- V体育2025版 - Actions
- VSports - Actions
- VSports在线直播 - Actions
- "VSports注册入口" Actions
- Actions
- Actions
- VSports app下载 - Actions
- Actions
- Actions
- Actions
- V体育官网入口 - Actions
- Actions
- Actions
- Actions
- "VSports最新版本" Actions
- Actions
- Actions
- Actions
- Actions
- "VSports" Actions
- "VSports在线直播" Actions
- Actions
- "VSports" Actions
- V体育2025版 - Actions
- Actions
- Actions
- Actions (V体育安卓版)
- Actions
- "V体育平台登录" Actions
- Actions
- V体育安卓版 - Actions
- "VSports手机版" Actions
- Actions
- Actions
- Actions (V体育平台登录)
- Actions
- Actions (V体育安卓版)
- "V体育2025版" Actions
- VSports最新版本 - Actions
- Actions (V体育平台登录)
- Actions
- Actions (VSports最新版本)
- V体育2025版 - Actions
- VSports手机版 - Actions
- Actions (VSports在线直播)
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- Actions
- Actions
- "V体育平台登录" Actions
- Actions
- Actions (VSports在线直播)
- Actions (VSports注册入口)
- Actions
- V体育安卓版 - Actions
- Actions (VSports)
- Actions (V体育官网入口)
- Actions (VSports)
- "VSports最新版本" Actions
- Actions (VSports app下载)
- Actions (V体育ios版)
- Actions
- Actions
- Actions
- VSports注册入口 - Actions
- Actions (V体育官网)
- Actions
- "VSports在线直播" Actions
- Actions
- Actions
- Actions (VSports注册入口)
- "V体育官网" Actions
- Actions (V体育安卓版)
- Actions
- VSports注册入口 - Actions
- Actions (VSports最新版本)
- "V体育官网" Actions
- Actions
- Actions
- "VSports app下载" Actions
- Actions (V体育官网入口)
- "VSports app下载" Actions
- Actions
- VSports app下载 - Actions
- "VSports最新版本" Actions
- "VSports" Actions
- Actions
- Actions
- Actions
- "V体育平台登录" Actions
- "VSports" Actions
- "V体育官网入口" Actions
- Actions
- "VSports在线直播" Actions
- V体育ios版 - Actions
- Actions (VSports)
- Actions
- "V体育2025版" Actions
- V体育安卓版 - Actions
- Actions
- VSports在线直播 - Actions
- Actions
- "VSports最新版本" Actions
- "VSports最新版本" Actions
- VSports最新版本 - Actions
- Actions
- Actions
- Actions
- Actions
- "V体育官网入口" Actions
- V体育2025版 - Actions
- "V体育官网入口" Actions
- "VSports注册入口" Actions
- Actions
- "V体育平台登录" Actions
- Actions
- Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- "VSports注册入口" Actions
- V体育官网 - Actions
- V体育2025版 - Actions
- "VSports app下载" Actions
- Actions (V体育官网)
- "VSports" Actions
- Actions
- Actions
- Actions
- V体育ios版 - Actions
- Actions (VSports app下载)
- Actions
- Actions (V体育安卓版)
- Actions
- Actions (V体育2025版)
- VSports注册入口 - Actions
- "VSports最新版本" Actions
- "V体育官网入口" Actions
- "V体育平台登录" Actions
- Actions (VSports手机版)
- Actions
- "VSports手机版" Actions
- V体育平台登录 - Actions
- Actions (VSports最新版本)
- Actions (V体育ios版)
- Actions
- VSports手机版 - Actions
- VSports注册入口 - Actions
- VSports - Actions
- Actions
- Actions (V体育2025版)
- Actions
- "V体育2025版" Actions
- Actions (VSports)
- "V体育安卓版" Actions
- Actions
- "VSports注册入口" Actions
- Actions
- "VSports" Actions
- "V体育平台登录" Actions
- "V体育ios版" Actions
- V体育平台登录 - Actions
- Actions
- "VSports注册入口" Actions
- Actions (V体育安卓版)
- Actions
- "V体育平台登录" Actions
- Actions
- VSports注册入口 - Actions
- VSports在线直播 - Actions
- Actions
- Actions
- "V体育安卓版" Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- "VSports在线直播" Actions
- Actions
- Actions (VSports)
- VSports注册入口 - Actions
- "V体育官网入口" Actions
- VSports - Actions
- Actions (VSports在线直播)
- Actions
- "V体育2025版" Actions
- Actions
- Actions (V体育ios版)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases (V体育官网入口)
Miscellaneous

