Integration of text- and data-mining using ontologies successfully selects disease gene candidates
- PMID: 15767279
- PMCID: VSports - PMC1065256
- DOI: 10.1093/nar/gki296
Integration of text- and data-mining using ontologies successfully selects disease gene candidates
VSports - Abstract
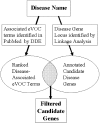
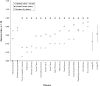
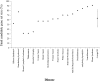
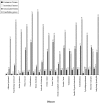
Genome-wide techniques such as microarray analysis, Serial Analysis of Gene Expression (SAGE), Massively Parallel Signature Sequencing (MPSS), linkage analysis and association studies are used extensively in the search for genes that cause diseases, and often identify many hundreds of candidate disease genes. Selection of the most probable of these candidate disease genes for further empirical analysis is a significant challenge. Additionally, identifying the genes that cause complex diseases is problematic due to low penetrance of multiple contributing genes. Here, we describe a novel bioinformatic approach that selects candidate disease genes according to their expression profiles. We use the eVOC anatomical ontology to integrate text-mining of biomedical literature and data-mining of available human gene expression data. To demonstrate that our method is successful and widely applicable, we apply it to a database of 417 candidate genes containing 17 known disease genes. We successfully select the known disease gene for 15 out of 17 diseases and reduce the candidate gene set to 63. 3% (+/-18. 8%) of its original size VSports手机版. This approach facilitates direct association between genomic data describing gene expression and information from biomedical texts describing disease phenotype, and successfully prioritizes candidate genes according to their expression in disease-affected tissues. .
Figures

References
-
- Pritchard J.K., Cox N.J. The allelic architecture of human disease genes: common disease-common variant… or not? Hum. Mol. Genet. 2002;11:2417–2423. - PubMed (V体育平台登录)
-
- Hoh J., Ott J. Genetic dissection of diseases: design and methods. Curr. Opin. Genet. Dev. 2004;14:229–232. - PubMed
-
- Glazier A.M., Nadeau J.H., Aitman T.J. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. - PubMed
-
- Tabor H.K., Risch N.J., Myers R.M. Opinion: candidate-gene approaches for studying complex genetic traits: practical considerations. Nature Rev. Genet. 2002;3:391–397. - PubMed
-
- Risch N.J. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. - "VSports注册入口" PubMed
Publication types
- "VSports手机版" Actions
MeSH terms
- Actions (V体育平台登录)
- "V体育安卓版" Actions
Grants and funding
"V体育官网" LinkOut - more resources
"V体育ios版" Full Text Sources
"V体育官网" Other Literature Sources

