Reovirus nonstructural protein mu NS recruits viral core surface proteins and entering core particles to factory-like inclusions
- PMID: 14747553
- PMCID: PMC369481
- DOI: 10.1128/jvi.78.4.1882-1892.2004
Reovirus nonstructural protein mu NS recruits viral core surface proteins and entering core particles to factory-like inclusions (V体育官网)
V体育官网入口 - Abstract
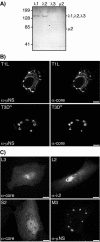
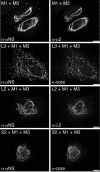
Mammalian reoviruses are thought to assemble and replicate within cytoplasmic, nonmembranous structures called viral factories. The viral nonstructural protein mu NS forms factory-like globular inclusions when expressed in the absence of other viral proteins and binds to the surfaces of the viral core particles in vitro. Given these previous observations, we hypothesized that one or more of the core surface proteins may be recruited to viral factories through specific associations with mu NS. We found that all three of these proteins--lambda 1, lambda 2, and sigma 2--localized to factories in infected cells but were diffusely distributed through the cytoplasm and nucleus when each was separately expressed in the absence of other viral proteins. When separately coexpressed with mu NS, on the other hand, each core surface protein colocalized with mu NS in globular inclusions, supporting the initial hypothesis VSports手机版. We also found that lambda 1, lambda 2, and sigma 2 each localized to filamentous inclusions formed upon the coexpression of mu NS and mu 2, a structurally minor core protein that associates with microtubules. The first 40 residues of mu NS, which are required for association with mu 2 and the RNA-binding nonstructural protein sigma NS, were not required for association with any of the three core surface proteins. When coexpressed with mu 2 in the absence of mu NS, each of the core surface proteins was diffusely distributed and displayed only sporadic, weak associations with mu 2 on filaments. Many of the core particles that entered the cytoplasm of cycloheximide-treated cells following entry and partial uncoating were recruited to inclusions of mu NS that had been preformed in those cells, providing evidence that mu NS can bind to the surfaces of cores in vivo. These findings expand a model for how viral and cellular components are recruited to the viral factories in infected cells and provide further evidence for the central but distinct roles of viral proteins mu NS and mu 2 in this process. .
Figures





References
-
- Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. - PubMed
-
- Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. - VSports app下载 - PMC - PubMed
-
- Borsa, J., and A. F. Graham. 1968. Reovirus: RNA polymerase activity in purified virions. Biochem. Biophys. Res. Commun. 33:895-901. - PubMed
Publication types
- V体育官网入口 - Actions
MeSH terms
- V体育平台登录 - Actions
- VSports手机版 - Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- "V体育安卓版" Actions
"VSports" Substances
- V体育ios版 - Actions
- "V体育官网入口" Actions
- "V体育2025版" Actions
- Actions (V体育2025版)
Grants and funding (V体育安卓版)
LinkOut - more resources
Full Text Sources
V体育平台登录 - Other Literature Sources
Research Materials
Miscellaneous

