Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein
- PMID: 14732903
- PMCID: PMC1181922
- DOI: 10.1086/381653
Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein
Abstract
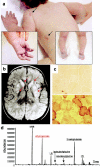
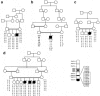
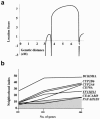
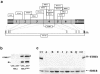
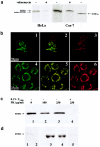
Ethylmalonic encephalopathy (EE) is a devastating infantile metabolic disorder affecting the brain, gastrointestinal tract, and peripheral vessels. High levels of ethylmalonic acid are detected in the body fluids, and cytochrome c oxidase activity is decreased in skeletal muscle. By use of a combination of homozygosity mapping, integration of physical and functional genomic data sets, and mutational screening, we identified GenBank D83198 as the gene responsible for EE. We also demonstrated that the D83198 protein product is targeted to mitochondria and internalized into the matrix after energy-dependent cleavage of a short leader peptide. The gene had previously been known as "HSCO" (for hepatoma subtracted clone one). However, given its role in EE, the name of the gene has been changed to "ETHE1 VSports手机版. " The severe consequences of its malfunctioning indicate an important role of the ETHE1 gene product in mitochondrial homeostasis and energy metabolism. .
"VSports注册入口" Figures

References (VSports最新版本)
Electronic-Database Information
-
- CELERA, http://www.celera.com
-
- Center for Genome Research, http://www.genome.wi.mit.edu/mpg/isfc/ (for neighborhood index tables)
References
-
- Aravind L (1999) An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol 1:69–91 - PubMed
-
- Attardi G, Schatz G (1988) Biogenesis of mitochondria. Annu Rev Cell Biol 4:289–333 - V体育官网 - PubMed
-
- Burlina AB, Dionisi-Vici C, Bennet MJ, Gibson KM, Servidei S, Bertini E, Hale DE, Schimidt-Sommerfeld E, Sabetta G, Zacchello F, Rinaldo P (1994) A new syndrome with ethylmalonic aciduria and normal fatty acid oxidation in fibroblasts. J Pediatr 124:79–86 - PubMed
V体育平台登录 - Publication types
MeSH terms (VSports注册入口)
- "V体育平台登录" Actions
- VSports - Actions
- Actions (V体育2025版)
- Actions (V体育官网入口)
- V体育ios版 - Actions
- Actions (V体育ios版)
- Actions (VSports app下载)
- Actions (V体育官网入口)
- "VSports app下载" Actions
- "VSports" Actions
- Actions (VSports手机版)
Substances (V体育安卓版)
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- Actions (VSports)
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

