The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses
- PMID: 14606959
- PMCID: PMC280656
- DOI: 10.1186/1471-2407-3-30
The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses
Abstract
Background: Histone deacetylase inhibitors (HDACIs) induce hyperacetylation of core histones modulating chromatin structure and affecting gene expression. These compounds are also able to induce growth arrest, cell differentiation, and apoptotic cell death of tumor cells in vitro as well as in vivo. Even though several genes modulated by HDAC inhibition have been identified, those genes clearly responsible for the biological effects of these drugs have remained elusive. We investigated the pharmacological effect of the HDACI and potential anti-cancer agent Trichostatin A (TSA) on primary T cells. VSports手机版.
Methods: To ascertain the effect of TSA on resting and activated T cells we used a model system where an enriched cell population consisting of primary T-cells was stimulated in vitro with immobilized anti-CD3/anti-CD28 antibodies whilst exposed to pharmacological concentrations of Trichostatin A V体育安卓版. .
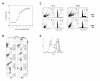
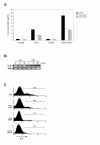
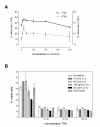
Results: We found that this drug causes a rapid decline in cytokine expression, accumulation of cells in the G1 phase of the cell cycle, and induces apoptotic cell death. The mitochondrial respiratory chain (MRC) plays a critical role in the apoptotic response to TSA, as dissipation of mitochondrial membrane potential and reactive oxygen species (ROS) scavengers block TSA-induced T-cell death. Treatment of T cells with TSA results in the altered expression of a subset of genes involved in T cell responses, as assessed by microarray gene expression profiling V体育ios版. We also observed up- as well as down-regulation of various costimulatory/adhesion molecules, such as CD28 and CD154, important for T-cell function. .
Conclusions: Taken together, our findings indicate that HDAC inhibitors have an immunomodulatory potential that may contribute to the potency and specificity of these antineoplastic compounds and might be useful in the treatment of autoimmune disorders VSports最新版本. .
Figures

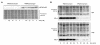

References
-
- Heinzel T, Lavinsky RM, Mullen TM, Söderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. - DOI - PubMed
MeSH terms
- Actions (VSports注册入口)
- "VSports" Actions
- "VSports手机版" Actions
- Actions (VSports手机版)
- "V体育安卓版" Actions
- V体育官网 - Actions
- VSports - Actions
- Actions (VSports在线直播)
- Actions (V体育安卓版)
- "VSports在线直播" Actions
- Actions (V体育平台登录)
- "VSports" Actions
- V体育平台登录 - Actions
Substances
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- "VSports注册入口" Actions
- "V体育安卓版" Actions
- V体育平台登录 - Actions
LinkOut - more resources (VSports最新版本)
Full Text Sources
Other Literature Sources
Research Materials

