Reversal of DNA alkylation damage by two human dioxygenases
- PMID: 12486230
- PMCID: "V体育2025版" PMC139200
- DOI: 10.1073/pnas.262589799
Reversal of DNA alkylation damage by two human dioxygenases (V体育ios版)
Abstract
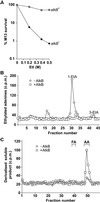
The Escherichia coli AlkB protein protects against the cytotoxicity of methylating agents by repair of the DNA lesions 1-methyladenine and 3-methylcytosine, which are generated in single-stranded stretches of DNA. AlkB is an alpha-ketoglutarate- and Fe(II)-dependent dioxygenase that oxidizes the relevant methyl groups and releases them as formaldehyde. Here, we identify two human AlkB homologs, ABH2 and ABH3, by sequence and fold similarity, functional assays, and complementation of the E. coli alkB mutant phenotype. The levels of their mRNAs do not appear to correlate with cell proliferation but tissue distributions are different. Both enzymes remove 1-methyladenine and 3-methylcytosine from methylated polynucleotides in an alpha-ketoglutarate-dependent reaction, and act by direct damage reversal with the regeneration of the unsubstituted bases. AlkB, ABH2, and ABH3 can also repair 1-ethyladenine residues in DNA with the release of acetaldehyde. VSports手机版.
Figures (VSports最新版本)



References
-
- Bodell W. J. & Singer, B. (1979) Biochemistry 18, 2860-2863. - PubMed
-
- Boiteux S. & Laval, J. (1982) Biochimie 64, 637-641. - PubMed
-
- Sedgwick, B. & Lindahl, T. (2002) Oncogene Rev., in press. - "VSports在线直播" PubMed
MeSH terms
- "VSports最新版本" Actions
- V体育官网入口 - Actions
- V体育安卓版 - Actions
- "V体育安卓版" Actions
- V体育2025版 - Actions
- Actions (V体育平台登录)
- "V体育官网入口" Actions
- V体育安卓版 - Actions
Substances
- "V体育官网入口" Actions
- Actions (V体育官网入口)
- Actions (V体育平台登录)
- VSports最新版本 - Actions
- VSports - Actions
- "V体育官网入口" Actions
LinkOut - more resources
Full Text Sources
V体育安卓版 - Other Literature Sources
Molecular Biology Databases

