Identification of a new class of cytochrome P450 from a Rhodococcus sp (V体育官网入口)
- PMID: 12081961
- PMCID: PMC135161
- DOI: 10.1128/JB.184.14.3898-3908.2002
Identification of a new class of cytochrome P450 from a Rhodococcus sp
Abstract
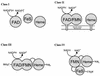
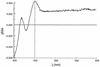
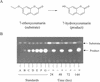
A degenerate set of PCR primers were used to clone a gene encoding a cytochrome P450 (the P450RhF gene) from Rhodococcus sp. strain NCIMB 9784 which is of unique primary structural organization. Surprisingly, analysis of the translation product revealed that the P450 is fused to a reductase domain at the C terminus which displays sequence conservation for dioxygenase reductase proteins. The reductase partner comprises flavin mononucleotide- and NADH-binding motifs and a [2Fe2S] ferredoxin-like center. The gene was engineered for heterologous expression in Escherichia coli, and conditions were found in which the enzyme was produced in a soluble form. A recombinant strain of E. coli was able to mediate the O dealkylation of 7-ethoxycoumarin in good yield, despite the absence of any recombinant redox proteins. This unprecedented finding leads us to propose that P450RhF represents the first example of a new class of cytochromes P450 in which the reducing equivalents are supplied by a novel reductase in a fused arrangement. VSports手机版.
Figures






References
-
- Ahn, K. S., and R. G. Wake. 1991. Variations and coding features of the sequence spanning the replication terminus of Bacillus subtilis 168 and W23 chromosomes. Gene 98:107-112. - "VSports在线直播" PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - "VSports在线直播" PubMed
-
- Anderson, J. F., and C. R. Hutchinson. 1992. Characterization of Saccharopolyspora erythraea cytochrome P450 genes and enzymes, including 6-deoxyerythronolide B hydroxylase. J. Bacteriol. 174:725-735. - PMC (VSports最新版本) - PubMed
-
- Black, S. M., J. A. Harikrishna, G. D. Szklarz, and W. L. Miller. 1994. The mitochondrial environment is required for activity of the cholesterol side-chain cleavage enzyme, cytochrome P450scc. Proc. Natl. Acad. Sci. USA 91:7247-7251. - VSports手机版 - PMC - PubMed
-
- Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K12. Science 277:1453-1474. - PubMed
Publication types
V体育2025版 - MeSH terms
- V体育安卓版 - Actions
- "V体育ios版" Actions
- "V体育平台登录" Actions
- "VSports手机版" Actions
- "V体育官网入口" Actions
Substances
Grants and funding (VSports最新版本)
"VSports在线直播" LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

