"VSports注册入口" Gene expression patterns in human liver cancers
- PMID: 12058060
- PMCID: PMC117615
- DOI: V体育ios版 - 10.1091/mbc.02-02-0023
Gene expression patterns in human liver cancers
Abstract
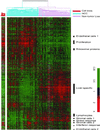
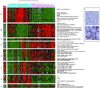
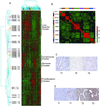
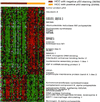
Hepatocellular carcinoma (HCC) is a leading cause of death worldwide. Using cDNA microarrays to characterize patterns of gene expression in HCC, we found consistent differences between the expression patterns in HCC compared with those seen in nontumor liver tissues. The expression patterns in HCC were also readily distinguished from those associated with tumors metastatic to liver. The global gene expression patterns intrinsic to each tumor were sufficiently distinctive that multiple tumor nodules from the same patient could usually be recognized and distinguished from all the others in the large sample set on the basis of their gene expression patterns alone VSports手机版. The distinctive gene expression patterns are characteristic of the tumors and not the patient; the expression programs seen in clonally independent tumor nodules in the same patient were no more similar than those in tumors from different patients. Moreover, clonally related tumor masses that showed distinct expression profiles were also distinguished by genotypic differences. Some features of the gene expression patterns were associated with specific phenotypic and genotypic characteristics of the tumors, including growth rate, vascular invasion, and p53 overexpression. .
Figures
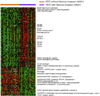
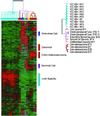
References
-
- Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Arii S, Mise M, Harada T, Furutani M, Ishigami S, Niwano M, Mizumoto M, Fukumoto M, Imamura M. Overexpression of matrix metalloproteinase 9 gene in hepatocellular carcinoma with invasive potential. Hepatology. 1996;24:316–322. - PubMed
-
- Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. - PubMed
-
- Caruso ML, Valentini AM. Overexpression of p53 in a large series of patients with hepatocellular carcinoma: a clinicopathological correlation. Anticancer Res. 1999;19:3853–3856. - PubMed (VSports在线直播)
-
- Cho RJ, Huang M, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ, Campbell MJ. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. - PubMed (V体育官网)
Publication types
- Actions (VSports注册入口)
VSports手机版 - MeSH terms
- V体育官网入口 - Actions
- Actions (VSports手机版)
- Actions (VSports在线直播)
- "VSports" Actions
- V体育平台登录 - Actions
- V体育平台登录 - Actions
- "V体育平台登录" Actions
- VSports app下载 - Actions
- "VSports在线直播" Actions
- Actions (V体育官网)
Substances (V体育ios版)
- Actions (V体育官网入口)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
"V体育平台登录" Miscellaneous

