V体育官网 - miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs
- PMID: 11914277
- PMCID: PMC155365 (V体育2025版)
- DOI: 10.1101/gad.974702
miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs
Abstract
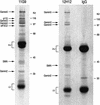
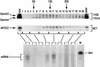
Gemin3 is a DEAD-box RNA helicase that binds to the Survival of Motor Neurons (SMN) protein and is a component of the SMN complex, which also comprises SMN, Gemin2, Gemin4, Gemin5, and Gemin6. Reduction in SMN protein results in Spinal muscular atrophy (SMA), a common neurodegenerative disease. The SMN complex has critical functions in the assembly/restructuring of diverse ribonucleoprotein (RNP) complexes. Here we report that Gemin3 and Gemin4 are also in a separate complex that contains eIF2C2, a member of the Argonaute protein family. This novel complex is a large approximately 15S RNP that contains numerous microRNAs (miRNAs). We describe 40 miRNAs, a few of which are identical to recently described human miRNAs, a class of small endogenous RNAs. The genomic sequences predict that miRNAs are likely to be derived from larger precursors that have the capacity to form stem-loop structures VSports手机版. .
Figures






References
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. - PubMed
-
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - "VSports注册入口" PubMed
-
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. - "V体育官网" PubMed
"VSports app下载" Publication types
- "VSports手机版" Actions
- "VSports最新版本" Actions
MeSH terms
- "V体育ios版" Actions
- VSports app下载 - Actions
- V体育官网入口 - Actions
- Actions (VSports最新版本)
- "VSports最新版本" Actions
- "VSports app下载" Actions
- V体育ios版 - Actions
- Actions (V体育2025版)
- "VSports" Actions
- "V体育ios版" Actions
- "V体育安卓版" Actions
- "VSports在线直播" Actions
- "VSports app下载" Actions
- VSports注册入口 - Actions
- "V体育官网入口" Actions
Substances
- "VSports在线直播" Actions
- "VSports最新版本" Actions
- "VSports app下载" Actions
- V体育安卓版 - Actions
- Actions (V体育ios版)
- Actions (V体育官网)
- "V体育平台登录" Actions
- "VSports" Actions
- V体育平台登录 - Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- "V体育ios版" Actions
- Actions (V体育安卓版)
LinkOut - more resources
"VSports手机版" Full Text Sources
Other Literature Sources
Molecular Biology Databases
