"VSports注册入口" PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2
- PMID: 11844753
- PMCID: V体育ios版 - PMC134856
- DOI: "VSports最新版本" 10.1128/JB.184.5.1253-1261.2002
PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2
Abstract
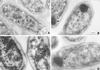
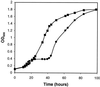
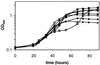
Salmonella enterica forms polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. These organelles are thought to consist of a proteinaceous shell that encases coenzyme B(12)-dependent diol dehydratase and perhaps other enzymes involved in 1,2-propanediol degradation. The function of these organelles is unknown, and no detailed studies of their structure have been reported. Genes needed for organelle formation and for 1,2-propanediol degradation are located at the 1,2-propanediol utilization (pdu) locus, but the specific genes involved in organelle formation have not been identified. Here, we show that the pduA gene encodes a shell protein required for the formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. A His(6)-PduA fusion protein was purified from a recombinant Escherichia coli strain and used for the preparation of polyclonal antibodies. The anti-PduA antibodies obtained were partially purified by a subtraction procedure and used to demonstrate that the PduA protein localized to the shell of the polyhedral organelles. In addition, electron microscopy studies established that strains with nonpolar pduA mutations were unable to form organelles. These results show that the pduA gene is essential for organelle formation and indicate that the PduA protein is a structural component of the shell of these organelles. Physiological studies of nonpolar pduA mutants were also conducted. Such mutants grew similarly to the wild-type strain at low concentrations of 1,2-propanediol but exhibited a period of interrupted growth in the presence of higher concentrations of this growth substrate. Growth tests also showed that a nonpolar pduA deletion mutant grew faster than the wild-type strain at low vitamin B(12) concentrations VSports手机版. These results suggest that the polyhedral organelles formed by S. enterica during growth on 1,2-propanediol are not involved in the concentration of 1,2-propanediol or coenzyme B(12), but are consistent with the hypothesis that these organelles moderate aldehyde production to minimize toxicity. .
Figures

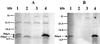

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Baker, S. H., S. C. Lorbach, M. Rodriguez-Buey, D. S. Williams, H. C. Aldrich, and J. M. Shively. 1999. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch. Microbiol. 172:233-239. - V体育官网 - PubMed
-
- Baker, S. H., D. S. Williams, H. C. Aldrich, A. C. Gambrell, and J. M. Shively. 2000. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch. Microbiol. 173:278-283. - V体育安卓版 - PubMed
Publication types
MeSH terms
- V体育平台登录 - Actions
- VSports注册入口 - Actions
- Actions (V体育2025版)
- "V体育平台登录" Actions
- Actions (V体育ios版)
- VSports手机版 - Actions
- VSports最新版本 - Actions
- "VSports" Actions
- Actions (V体育平台登录)
Substances (VSports注册入口)
- V体育官网 - Actions
- V体育官网入口 - Actions
- "V体育ios版" Actions
- V体育官网入口 - Actions
Grants and funding
LinkOut - more resources
"VSports注册入口" Full Text Sources
Other Literature Sources
Molecular Biology Databases (VSports注册入口)

