FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity
- PMID: 11641274
- PMCID: PMC312814
- DOI: "V体育平台登录" 10.1101/gad.924501
V体育安卓版 - FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity
Erratum in
-
Corrigendum: FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity.Genes Dev. 2025 Jul 1;39(13-14):907. doi: 10.1101/gad.352958.125. Genes Dev. 2025. PMID: 40592560 Free PMC article. No abstract available.
Abstract (VSports)
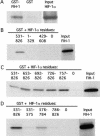
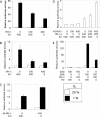
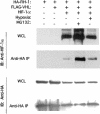
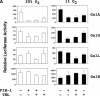
Hypoxia-inducible factor 1 (HIF-1) is a master regulator of oxygen homeostasis that controls angiogenesis, erythropoiesis, and glycolysis via transcriptional activation of target genes under hypoxic conditions VSports手机版. O(2)-dependent binding of the von Hippel-Lindau (VHL) tumor suppressor protein targets the HIF-1alpha subunit for ubiquitination and proteasomal degradation. The activity of the HIF-1alpha transactivation domains is also O(2) regulated by a previously undefined mechanism. Here, we report the identification of factor inhibiting HIF-1 (FIH-1), a protein that binds to HIF-1alpha and inhibits its transactivation function. In addition, we demonstrate that FIH-1 binds to VHL and that VHL also functions as a transcriptional corepressor that inhibits HIF-1alpha transactivation function by recruiting histone deacetylases. Involvement of VHL in association with FIH-1 provides a unifying mechanism for the modulation of HIF-1alpha protein stabilization and transcriptional activation in response to changes in cellular O(2) concentration. .
Figures


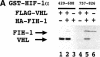
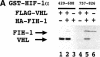
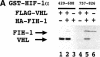
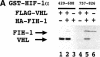





References
-
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature. 1998;392:405–408. - PubMed
-
- Bigner SH, Vogelstein B. Cytogenetics and molecular genetics of malignant gliomas and medulloblastoma. Brain Pathol. 1990;1:12–18. - PubMed
-
- Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci. 2000;97:9082–9087. - "V体育官网入口" PMC - PubMed
-
- Carmeliet P, Dor Y, Herbert J-M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation, and tumour angiogenesis. Nature. 1998;394:485–490. - PubMed
Publication types
VSports - MeSH terms
- Actions (VSports手机版)
- VSports手机版 - Actions
- VSports在线直播 - Actions
- V体育官网入口 - Actions
- Actions (VSports手机版)
- "VSports app下载" Actions
- "V体育2025版" Actions
- Actions (V体育官网入口)
- Actions (V体育ios版)
- Actions (V体育2025版)
- Actions (VSports)
- Actions (V体育平台登录)
- "V体育官网" Actions
- VSports在线直播 - Actions
- Actions (V体育官网)
- Actions (VSports在线直播)
- "V体育ios版" Actions
- "VSports最新版本" Actions
"V体育官网" Substances
- Actions (V体育2025版)
- V体育官网 - Actions
- "V体育平台登录" Actions
- Actions (VSports手机版)
- Actions (V体育ios版)
- V体育ios版 - Actions
- Actions (V体育2025版)
- Actions (V体育2025版)
- V体育官网入口 - Actions
- Actions (V体育安卓版)
- "V体育平台登录" Actions
V体育安卓版 - Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
