Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene
- PMID: 11514597
- PMCID: PMC2193506
- DOI: 10.1084/jem.194.4.393
Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene
VSports手机版 - Abstract
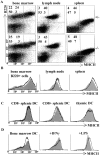
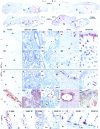
MHC class II (MHCII) molecules play a pivotal role in the induction and regulation of immune responses. The transcriptional coactivator class II transactivator (CIITA) controls MHCII expression. The CIITA gene is regulated by three independent promoters (pI, pIII, pIV). We have generated pIV knockout mice. These mice exhibit selective abrogation of interferon (IFN)-gamma-induced MHCII expression on a wide variety of non-bone marrow-derived cells, including endothelia, epithelia, astrocytes, and fibroblasts VSports手机版. Constitutive MHCII expression on cortical thymic epithelial cells, and thus positive selection of CD4(+) T cells, is also abolished. In contrast, constitutive and inducible MHCII expression is unaffected on professional antigen-presenting cells, including B cells, dendritic cells, and IFN-gamma-activated cells of the macrophage lineage. pIV(-/-) mice have thus allowed precise definition of CIITA pIV usage in vivo. Moreover, they represent a unique animal model for studying the significance and contribution of MHCII-mediated antigen presentation by nonprofessional antigen-presenting cells in health and disease. .
Figures










Comment in
-
The pIV-otal class II transactivator promoter regulates major histocompatibility complex class II expression in the thymus.J Exp Med. 2001 Aug 20;194(4):F15-8. doi: 10.1084/jem.194.4.f15. J Exp Med. 2001. PMID: 11514611 Free PMC article. No abstract available.
References
-
- Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. - PubMed
-
- Viret C., Janeway C.A. MHC and T cell development. Rev. Immunogenet. 1999;1:91–104. - PubMed
-
- Harton J.A., Ting J.P. Class II transactivatormastering the art of major histocompatibility complex expression. Mol. Cell Biol. 2000;20:6185–6194. - PMC (V体育2025版) - PubMed
-
- Boss J.M. Regulation of transcription of MHC class II genes. Curr. Opin. Immunol. 1997;9:107–113. - PubMed
-
- Reith W., Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 2001;19:331–373. - PubMed
"VSports" MeSH terms
- VSports手机版 - Actions
- "VSports app下载" Actions
- "VSports注册入口" Actions
- "VSports" Actions
- "VSports手机版" Actions
- "VSports注册入口" Actions
- VSports手机版 - Actions
- "VSports最新版本" Actions
Substances
- "VSports注册入口" Actions
- "V体育2025版" Actions
- "VSports手机版" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases (VSports最新版本)
Research Materials
Miscellaneous

