V体育官网入口 - Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA
- PMID: 11461998
- PMCID: PMC114961
- DOI: 10.1128/JVI.75.16.7252-7265.2001 (V体育ios版)
V体育平台登录 - Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA
Abstract
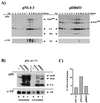
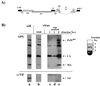
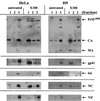
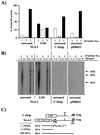
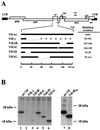
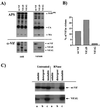
The human immunodeficiency virus type 1 (HIV-1) Vif protein plays a critical role in the production of infectious virions. Previous studies have demonstrated the presence of small amounts of Vif in virus particles. However, Vif packaging was assumed to be nonspecific, and its functional significance has been questioned. We now report that packaging of Vif is dependent on the packaging of viral genomic RNA in both permissive and restrictive HIV-1 target cells. Mutations in the nucleocapsid zinc finger domains that abrogate packaging of viral genomic RNA abolished packaging of Vif. Additionally, an RNA packaging-defective virus exhibited significantly reduced packaging of Vif. Finally, deletion of a putative RNA-interacting domain in Vif abolished packaging of Vif into virions. Virion-associated Vif was resistant to detergent extraction and copurified with components of the viral nucleoprotein complex and functional reverse transcription complexes. Thus, Vif is specifically packaged into virions as a component of the viral nucleoprotein complex VSports手机版. Our data suggest that the specific association of Vif with the viral nucleoprotein complex might be functionally significant and could be a critical requirement for infectivity of viruses produced from restrictive host cells. .
Figures

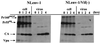

References
-
- Akari H, Sakuragi J, Takebe Y, Tomonaga K, Kawamura M, Fukasawa M, Miura T, Shinjo T, Hayami M. Biological characterization of human immunodeficiency virus type 1 and type 2 mutants in human peripheral blood mononuclear cells. Arch Virol. 1992;123:157–167. - PubMed
-
- Akari H, Uchiyama T, Fukumori T, Iida S, Koyama A H, Adachi A. Pseudotyping human immunodeficiency virus type 1 by vesicular stomatitis virus G protein does not reduce the cell-dependent requirement of vif for optimal infectivity: functional difference between Vif and Nef. J Gen Virol. 1999;80:2945–2949. - PubMed
-
- Bess J W, Jr, Gorelick R J, Bosche W J, Henderson L E, Arthur L O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. - PubMed (V体育安卓版)
VSports最新版本 - Publication types
MeSH terms (VSports)
- V体育安卓版 - Actions
- V体育安卓版 - Actions
- "VSports最新版本" Actions
- Actions (V体育安卓版)
- V体育ios版 - Actions
Substances
- Actions (V体育官网入口)
- Actions (VSports app下载)
Grants and funding
LinkOut - more resources (V体育平台登录)
Full Text Sources
Other Literature Sources

