Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine
- PMID: 11342591
- PMCID: "V体育安卓版" PMC2193429
- DOI: 10.1084/jem.193.9.1067
Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine
"VSports在线直播" Abstract
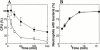
Defensins, antimicrobial peptides of the innate immune system, protect human mucosal epithelia and skin against microbial infections and are produced in large amounts by neutrophils. The bacterial pathogen Staphylococcus aureus is insensitive to defensins by virtue of an unknown resistance mechanism. We describe a novel staphylococcal gene, mprF, which determines resistance to several host defense peptides such as defensins and protegrins. An mprF mutant strain was killed considerably faster by human neutrophils and exhibited attenuated virulence in mice, indicating a key role for defensin resistance in the pathogenicity of S. aureus. Analysis of membrane lipids demonstrated that the mprF mutant no longer modifies phosphatidylglycerol with l-lysine VSports手机版. As this unusual modification leads to a reduced negative charge of the membrane surface, MprF-mediated peptide resistance is most likely based on repulsion of the cationic peptides. Accordingly, inactivation of mprF led to increased binding of antimicrobial peptides by the bacteria. MprF has no similarity with genes of known function, but related genes were identified in the genomes of several pathogens including Mycobacterium tuberculosis, Pseudomonas aeruginosa, and Enterococcus faecalis. MprF thus constitutes a novel virulence factor, which may be of general relevance for bacterial pathogens and represents a new target for attacking multidrug resistant bacteria. .
Figures (VSports最新版本)


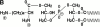
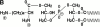
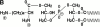


References
-
- Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. - PubMed
-
- Sieradzki K., Roberts R.B., Haber S.W., Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 1999;340:517–523. - VSports最新版本 - PubMed
-
- Peschel A., Otto M., Jack R.W., Kalbacher H., Jung G., Götz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins and other antimicrobial peptides. J. Biol. Chem. 1999;274:8405–8410. - PubMed (VSports最新版本)
-
- Harder J., Bartels J., Christophers E., Schröder J.-M. A peptide antibiotic from human skin. Nature. 1997;387:861. - V体育2025版 - PubMed
-
- Lehrer R.I., Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999;11:23–27. - PubMed
"V体育官网" Publication types
- Actions (VSports app下载)
MeSH terms (VSports最新版本)
- "VSports" Actions
- "V体育安卓版" Actions
- VSports app下载 - Actions
- V体育官网入口 - Actions
- VSports - Actions
- VSports - Actions
- VSports在线直播 - Actions
- Actions (VSports在线直播)
- VSports手机版 - Actions
- "VSports注册入口" Actions
V体育官网入口 - Substances
- V体育平台登录 - Actions
- V体育官网 - Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- VSports注册入口 - Actions
- Actions (V体育2025版)
- "VSports" Actions
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育平台登录 - Medical
Molecular Biology Databases
Research Materials

