"VSports手机版" Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains
- PMID: 11226167
- PMCID: PMC140198
- DOI: 10.1093/emboj/20.1.165 (V体育官网)
"V体育2025版" Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains
Abstract
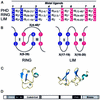
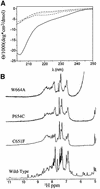
Plant homeodomain (PHD) domains are found in >400 eukaryotic proteins, many of which are transcriptional regulators VSports手机版. Naturally occurring point mutations or deletions of this domain contribute to a variety of human diseases, including ATRX syndrome, myeloid leukemias and autoimmune dysfunction. Here we report the first structural characterization of a PHD domain. Our studies reveal that the PHD domain from KAP-1 corepressor binds zinc in a cross-brace topology between anti-parallel ss-strands reminiscent of RING (really interesting new gene) domains. Using a mutational analysis, we define the structural features required for transcriptional repression by KAP-1 and explain naturally occurring, disease-causing mutations in PHD domains of other proteins. From a comparison of this PHD structure with previously reported RING and LIM (Lin11/Isl-1/Mec-3) structures, we infer sequence determinants that allow discrimination among PHD, RING and LIM motifs. .
Figures
References
-
- Aapola U. et al. (2000) Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. - "VSports在线直播" PubMed
-
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
-
- Barlow P.N., Luisi,B., Milner,A., Elliott,M. and Everett,R. (1994) Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol., 237, 201–211. - PubMed
-
- Bellon S.F., Rodgers,K.K., Schatz,D.G., Coleman,J.E. and Steitz,T.A. (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nature Struct. Biol., 4, 586–591. - PubMed
-
- Bochar D.A., Savard,J., Wang,W., Lafleur,D.W., Moore,P., Cote,J. and Shiekhattar,R. (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl Acad. Sci. USA, 97, 1038–1043. - "V体育官网" PMC - PubMed
"VSports app下载" Publication types
- "V体育安卓版" Actions
MeSH terms
- V体育官网入口 - Actions
- V体育平台登录 - Actions
- V体育官网入口 - Actions
- V体育官网 - Actions
- Actions (VSports注册入口)
- "V体育官网" Actions
- "VSports app下载" Actions
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions (VSports)
- "V体育平台登录" Actions
Substances
- "VSports app下载" Actions
V体育平台登录 - Grants and funding
V体育官网入口 - LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育官网入口)
"V体育安卓版" Molecular Biology Databases
Miscellaneous

