VSports手机版 - Predicting protein function by genomic context: quantitative evaluation and qualitative inferences
- PMID: 10958638
- PMCID: PMC310926 (V体育2025版)
- DOI: 10.1101/gr.10.8.1204
Predicting protein function by genomic context: quantitative evaluation and qualitative inferences
VSports手机版 - Abstract
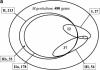
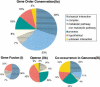
Various new methods have been proposed to predict functional interactions between proteins based on the genomic context of their genes. The types of genomic context that they use are Type I: the fusion of genes; Type II: the conservation of gene-order or co-occurrence of genes in potential operons; and Type III: the co-occurrence of genes across genomes (phylogenetic profiles). Here we compare these types for their coverage, their correlations with various types of functional interaction, and their overlap with homology-based function assignment. We apply the methods to Mycoplasma genitalium, the standard benchmarking genome in computational and experimental genomics. Quantitatively, conservation of gene order is the technique with the highest coverage, applying to 37% of the genes VSports手机版. By combining gene order conservation with gene fusion (6%), the co-occurrence of genes in operons in absence of gene order conservation (8%), and the co-occurrence of genes across genomes (11%), significant context information can be obtained for 50% of the genes (the categories overlap). Qualitatively, we observe that the functional interactions between genes are stronger as the requirements for physical neighborhood on the genome are more stringent, while the fraction of potential false positives decreases. Moreover, only in cases in which gene order is conserved in a substantial fraction of the genomes, in this case six out of twenty-five, does a single type of functional interaction (physical interaction) clearly dominate (>80%). In other cases, complementary function information from homology searches, which is available for most of the genes with significant genomic context, is essential to predict the type of interaction. Using a combination of genomic context and homology searches, new functional features can be predicted for 10% of M. genitalium genes. .
Figures


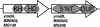
References
-
- Aravind L, Koonin E. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. - PubMed
-
- Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. Predicting function: From genes to genomes and back. J Mol Biol. 1998;283:707–725. - V体育官网入口 - PubMed
-
- Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. - PubMed
Publication types
MeSH terms (VSports手机版)
- V体育ios版 - Actions
- "VSports" Actions
- Actions (V体育官网入口)
- VSports在线直播 - Actions
- Actions (V体育安卓版)
Substances (VSports)
- "VSports" Actions
LinkOut - more resources
V体育ios版 - Full Text Sources
Other Literature Sources
