The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding (V体育平台登录)
- PMID: 10768965
- PMCID: "VSports" PMC97480
- DOI: 10.1128/IAI.68.5.2720-2727.2000
The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding
Abstract
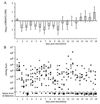
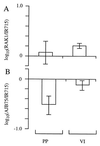
Little is known about factors which enable Salmonella serotypes to circulate within populations of livestock and domestic fowl VSports手机版. We have identified a DNA region which is present in Salmonella serotypes commonly isolated from livestock and domestic fowl (S. enterica subspecies I) but absent from reptile-associated Salmonella serotypes (S. bongori and S. enterica subspecies II to VII). This DNA region was cloned from Salmonella serotype Typhimurium and sequence analysis revealed the presence of a 6,105-bp open reading frame, designated shdA, whose product's deduced amino acid sequence displayed homology to that of AIDA-I from diarrheagenic Escherichia coli, MisL of serotype Typhimurium, and IcsA of Shigella flexneri. The shdA gene was located adjacent to xseA at 52 min, in a 30-kb DNA region which is not present in Escherichia coli K-12. A serotype Typhimurium shdA mutant was shed with the feces in reduced numbers and for a shorter period of time compared to its isogenic parent. A possible role for the shdA gene during the expansion in host range of S. enterica subspecies I to include warm-blooded vertebrates is discussed. .
Figures




References
-
- Aleksic S, Heinzerling F, Bockemühl J. Human infection caused by salmonellae of subspecies II to VI in Germany, 1977-1992. Zentbl Bakteriol. 1996;283:391–398. - V体育2025版 - PubMed
-
- Anderson R M. Evolutionary pressures in the spread and persistence of infectious agents in vertebrate populations. Parasitology. 1995;111(Suppl.):S15–S31. - PubMed
-
- Anderson R M, May R M. Coevolution of host and parasites. Parasitology. 1982;85:411–426. - PubMed
-
- Aserkoff B, Schroeder S A, Brachman P S. Salmonellosis in the United States—a five-year review. Am J Epidemiol. 1970;92:13–24. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1994.
Publication types
- Actions (VSports注册入口)
- Actions (V体育官网)
MeSH terms
- "V体育官网入口" Actions
- VSports手机版 - Actions
- Actions (VSports)
- VSports app下载 - Actions
- V体育2025版 - Actions
- VSports app下载 - Actions
- V体育2025版 - Actions
- "V体育平台登录" Actions
V体育平台登录 - Substances
- VSports - Actions
- "V体育2025版" Actions
- "V体育安卓版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

