"VSports在线直播" M-like proteins of Streptococcus dysgalactiae
- PMID: 10603401
- PMCID: PMC97134 (VSports手机版)
- DOI: 10.1128/IAI.68.1.294-302.2000
M-like proteins of Streptococcus dysgalactiae
Abstract
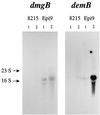
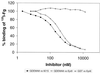
Streptococcus dysgalactiae is one of the most important bacterial species isolated from bovine mastitis. To identify potential virulence factors of this species we prepared chromosomal DNA from strain 8215 and constructed a phage display library. By affinity selection of the library against fibrinogen (Fg), we isolated and characterized a gene, called demA, encoding a protein with the molecular mass of approximately 58 kDa, called DemA, displaying both plasma protein binding properties and sequence similarities with the M and M-like proteins of other streptococcal species VSports手机版. Purified recombinant DemA protein was found to completely inhibit Fg-binding to cells of S. dysgalactiae. A continued sequence analysis revealed that the demA gene was preceded by an open reading frame (dmgA) coding for a putative protein, called DmgA, with high similarities to the Mga proteins of Streptococcus pyogenes. By additional cloning, the corresponding dmgA and demA genes from another strain, called Epi9, were isolated and analyzed. These genes, called dmgB and demB, respectively, revealed a high degree of similarity to the corresponding genes in strain 8215. Increased binding of Fg by cells of strain Epi9, grown in an atmosphere with 10% CO(2), was correlated to an enhanced transcription of the demB gene as shown in a Northern blot. Strain 8215 did not respond to CO(2), which could be explained by a nonfunctional dmgA gene due to insertion of an insertion sequence element. Based on sequence similarities of the described proteins to Mga, M, and M-like proteins and the response to elevated level of CO(2), we suggest that the dmg and dem genes are members of a regulon similar to the described mga regulon in S. pyogenes, which encodes several virulence factors in this species. .
Figures


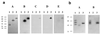

References
-
- Almeida R A, Oliver S P. Invasion of bovine mammary epithelial cells by Streptococcus dysgalactiae. J Dairy Sci. 1995;78:1310–1317. - PubMed
-
- Boyle M D. Variation of multifunctional surface binding proteins—a virulence strategy for group A streptococci? J Theor Biol. 1995;173:415–426. - PubMed
-
- Boyle M D, Raeder R, Flosdorff A, Podbielski A. Role of emm and mrp genes in the virulence of group A streptococcal isolate 64/14 in a mouse model of skin infection. J Infect Dis. 1998;177:991–997. - PubMed
-
- Calvinho L F, Almeida R A. Influence of Streptococcus dysgalactiae surface hydrophobicity on adherence to mammary epithelial cells and phagocytosis by mammary macrophages. Zentbl Vetmed Reihe B. 1996;43:257–266. - PubMed
-
- Calvinho L F, Oliver S P. Invasion and persistence of Streptococcus dysgalactiae within bovine mammary epithelial cells. J Dairy Sci. 1998;81:678–686. - V体育ios版 - PubMed
Publication types
- "VSports最新版本" Actions
MeSH terms (V体育官网入口)
- "VSports手机版" Actions
- "VSports手机版" Actions
- VSports注册入口 - Actions
- "VSports最新版本" Actions
- V体育平台登录 - Actions
- Actions (V体育ios版)
- "V体育ios版" Actions
- Actions (VSports手机版)
- Actions (VSports在线直播)
- VSports手机版 - Actions
- "VSports app下载" Actions
- VSports最新版本 - Actions
- Actions (V体育ios版)
- "V体育2025版" Actions
VSports注册入口 - Substances
- "V体育2025版" Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- Actions (VSports app下载)
Associated data
- Actions
- "V体育平台登录" Actions
"VSports app下载" LinkOut - more resources
"VSports手机版" Full Text Sources
"VSports最新版本" Other Literature Sources

