Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis
- PMID: 10541552
- PMCID: PMC317106
- DOI: 10.1101/gad.13.20.2658
Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis
VSports注册入口 - Abstract
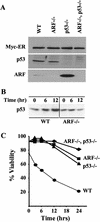
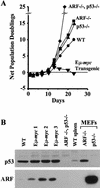
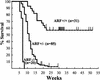
Transgenic mice expressing the c-Myc oncogene driven by the immunoglobulin heavy chain enhancer (Emu) develop B-cell lymphoma and exhibit a mean survival time of approximately 6 months. The protracted latent period before the onset of frank disease likely reflects the ability of c-Myc to induce a p53-dependent apoptotic program that initially protects animals against tumor formation but is disabled when overtly malignant cells emerge. In cultured primary mouse embryo fibroblasts, c-Myc activates the p19(ARF)-Mdm2-p53 tumor suppressor pathway, enhancing p53-dependent apoptosis but ultimately selecting for surviving immortalized cells that have sustained either p53 mutation or biallelic ARF deletion VSports手机版. Here we report that p53 and ARF also potentiate Myc-induced apoptosis in primary pre-B-cell cultures, and that spontaneous inactivation of the ARF-Mdm2-p53 pathway occurs frequently in tumors arising in Emu-myc transgenic mice. Many Emu-myc lymphomas sustained either p53 (28%) or ARF (24%) loss of function, whereas Mdm2 levels were elevated in others. Its overexpression in some tumors lacking p53 function raises the possibility that Mdm2 can contribute to lymphomagenesis by interacting with other targets. Emu-myc transgenic mice hemizygous for ARF displayed accelerated disease (11-week mean survival), and 80% of these tumors lost the wild-type ARF allele. All ARF-null Emu-myc mice died of lymphoma within a few weeks of birth. About half of the tumors arising in ARF hemizygous or ARF nullizygous Emu-myc transgenic mice also overexpressed Mdm2. Therefore, Myc activation strongly selects for spontaneous inactivation of the ARF-Mdm2-p53 pathway in vivo, cancelling its protective checkpoint function and accelerating progression to malignancy. .
Figures (VSports注册入口)


References
-
- Adams JM, Cory S. Oncogene cooperation in leukaemogenesis. Cancer Surveys. 1992;15:119–141. - PubMed
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Alitalo K, Koskinen P, Makela TP, Saksela K, Sistonen L, Winqvist R. Myc oncogenes: Activation and amplification. Biochim Biophys Acta. 1987;907:1–32. - PubMed
-
- Alkema MJ, Jacobs H, van Lohuizen M, Berns A. Perturbation of B and T cell development and predisposition to lymphomagenesis in Eμ-bmi-1 transgenic mice require the Bmi-1 RING finger. Oncogene. 1997;15:899–910. - VSports最新版本 - PubMed
-
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. - PubMed
Publication types (VSports手机版)
V体育ios版 - MeSH terms
- Actions (VSports)
- "V体育2025版" Actions
- "V体育官网" Actions
- V体育平台登录 - Actions
- "VSports手机版" Actions
- Actions (VSports注册入口)
- V体育官网入口 - Actions
- "VSports手机版" Actions
- Actions (V体育2025版)
- Actions (VSports在线直播)
- V体育官网入口 - Actions
- Actions (V体育官网入口)
- Actions (VSports注册入口)
- V体育官网 - Actions
Substances
- Actions (V体育安卓版)
- VSports手机版 - Actions
"VSports手机版" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials (VSports在线直播)
"V体育安卓版" Miscellaneous
