Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally
- PMID: 10409745
- PMCID: PMC84408
- DOI: 10.1128/MCB.19.8.5557
Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally
Abstract
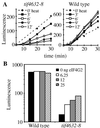
The initiation of translation in eukaryotes requires several multisubunit complexes, including eukaryotic translation initiation factor 4F (eIF4F). In higher eukaryotes eIF4F is composed of the cap binding protein eIF4E, the adapter protein eIF4G, and the RNA-stimulated ATPase eIF4A. The association of eIF4A with Saccharomyces cerevisiae eIF4F has not yet been demonstrated, and therefore the degree to which eIF4A's conserved function relies upon this association has remained unclear. Here we report an interaction between yeast eIF4G and eIF4A. Specifically, we found that the growth arrest phenotype associated with three temperature-sensitive alleles of yeast eIF4G2 was suppressed by excess eIF4A and that this suppression was allele specific. In addition, in vitro translation extracts derived from an eIF4G2 mutant strain could be heat inactivated, and this inactivation could be reversed upon the addition of recombinant eIF4A. Finally, in vitro binding between yeast eIF4G and eIF4A was demonstrated, as was diminished binding between mutant eIF4G2 proteins and eIF4A. In total, these data indicate that yeast eIF4G and eIF4A physically associate and that this association performs an essential function. VSports手机版.
Figures





References
-
- Boeke J D, Trueheart J, Natsoulis G, Fink G. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
-
- Chuang R Y, Weaver P L, Liu Z, Chang T H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1468–1471. - PubMed
Publication types
MeSH terms
- "V体育ios版" Actions
- "VSports最新版本" Actions
- "VSports注册入口" Actions
- Actions (V体育安卓版)
- "VSports最新版本" Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- V体育ios版 - Actions
- Actions (V体育2025版)
- Actions (V体育安卓版)
Substances
- V体育官网入口 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
V体育官网 - Miscellaneous
