A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors
- PMID: 10082585
- PMCID: PMC84112
- DOI: 10.1128/MCB.19.4.3184
A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors (VSports手机版)
Abstract
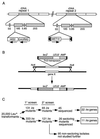
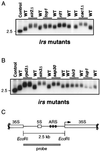
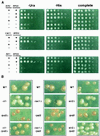
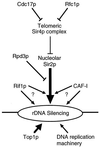
Transcriptional silencing in Saccharomyces cerevisiae occurs at several genetic loci, including the ribosomal DNA (rDNA). Silencing at telomeres (telomere position effect [TPE]) and the cryptic mating-type loci (HML and HMR) depends on the silent information regulator genes, SIR1, SIR2, SIR3, and SIR4. However, silencing of polymerase II-transcribed reporter genes integrated within the rDNA locus (rDNA silencing) requires only SIR2. The mechanism of rDNA silencing is therefore distinct from TPE and HM silencing. Few genes other than SIR2 have so far been linked to the rDNA silencing process. To identify additional non-Sir factors that affect rDNA silencing, we performed a genetic screen designed to isolate mutations which alter the expression of reporter genes integrated within the rDNA. We isolated two classes of mutants: those with a loss of rDNA silencing (lrs) phenotype and those with an increased rDNA silencing (irs) phenotype. Using transposon mutagenesis, lrs mutants were found in 11 different genes, and irs mutants were found in 22 different genes. Surprisingly, we did not isolate any genes involved in rRNA transcription. Instead, multiple genes associated with DNA replication and modulation of chromatin structure were isolated. We describe these two gene classes, and two previously uncharacterized genes, LRS4 and IRS4. Further characterization of the lrs and irs mutants revealed that many had alterations in rDNA chromatin structure. Several lrs mutants, including those in the cdc17 and rfc1 genes, caused lengthened telomeres, consistent with the hypothesis that telomere length modulates rDNA silencing VSports手机版. Mutations in the HDB (RPD3) histone deacetylase complex paradoxically increased rDNA silencing by a SIR2-dependent, SIR3-independent mechanism. Mutations in rpd3 also restored mating competence selectively to sir3Delta MATalpha strains, suggesting restoration of silencing at HMR in a sir3 mutant background. .
Figures (VSports注册入口)





References (VSports最新版本)
-
- Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Austriaco N R, Jr, Guarente L P. Changes in telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:9768–9772. - V体育2025版 - PMC - PubMed
-
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. - "VSports最新版本" PMC - PubMed
Publication types
- Actions (V体育2025版)
MeSH terms
- Actions (VSports手机版)
- "VSports最新版本" Actions
- Actions (V体育官网入口)
- "V体育平台登录" Actions
- "VSports最新版本" Actions
- Actions (V体育安卓版)
- "VSports在线直播" Actions
- VSports手机版 - Actions
- Actions (VSports最新版本)
- Actions (VSports手机版)
- V体育安卓版 - Actions
- "V体育官网" Actions
- Actions (V体育2025版)
- V体育官网入口 - Actions
V体育ios版 - Substances
- V体育官网入口 - Actions
- VSports最新版本 - Actions
- V体育官网 - Actions
- Actions (VSports app下载)
- Actions (V体育安卓版)
- VSports在线直播 - Actions
- "V体育平台登录" Actions
- V体育ios版 - Actions
V体育官网 - Grants and funding
"VSports" LinkOut - more resources
Full Text Sources
V体育官网入口 - Other Literature Sources
Molecular Biology Databases
